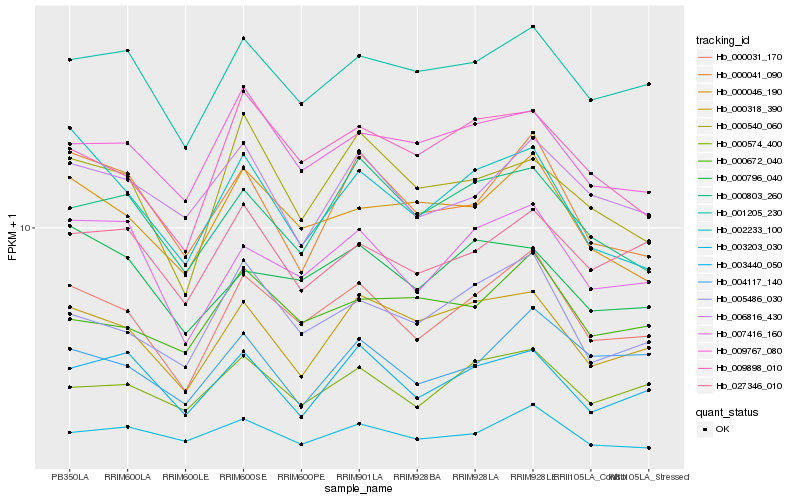
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_170 |
0.0 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007416_160 |
0.0754112755 |
- |
- |
PREDICTED: centromere-associated protein E isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_005486_030 |
0.0830826579 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 4 |
Hb_001205_230 |
0.0874933161 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 5 |
Hb_000046_190 |
0.0907318982 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 6 |
Hb_000574_400 |
0.091242906 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 7 |
Hb_000318_390 |
0.0917343935 |
- |
- |
PREDICTED: uncharacterized protein LOC105631693 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009898_010 |
0.0919864762 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 9 |
Hb_003203_030 |
0.0996244041 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 10 |
Hb_002233_100 |
0.0997836214 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 11 |
Hb_000041_090 |
0.1019003165 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 12 |
Hb_009767_080 |
0.1022025204 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 13 |
Hb_000803_260 |
0.1023005215 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 14 |
Hb_027346_010 |
0.1032190748 |
- |
- |
- |
| 15 |
Hb_000672_040 |
0.1042264832 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 16 |
Hb_006816_430 |
0.1048972141 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004117_140 |
0.1049735966 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 18 |
Hb_003440_050 |
0.1055887124 |
- |
- |
hypothetical protein JCGZ_05806 [Jatropha curcas] |
| 19 |
Hb_000540_060 |
0.1060352219 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 20 |
Hb_000796_040 |
0.1063808167 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |