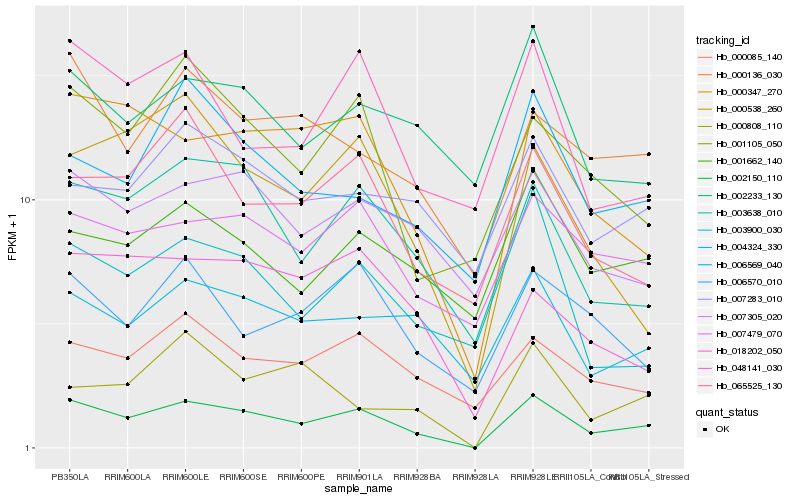
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_110 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 2 |
Hb_000347_270 |
0.1442267939 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 3 |
Hb_003638_010 |
0.1481560464 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 4 |
Hb_007479_070 |
0.1603072345 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 5 |
Hb_001105_050 |
0.1648173365 |
- |
- |
PREDICTED: uncharacterized protein LOC105642744 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000085_140 |
0.1650352668 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 7 |
Hb_048141_030 |
0.1660827596 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g59040-like [Prunus mume] |
| 8 |
Hb_000136_030 |
0.1663366348 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 9 |
Hb_065525_130 |
0.1664662997 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004324_330 |
0.1670798398 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 11 |
Hb_000538_260 |
0.1674422597 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 12 |
Hb_007305_020 |
0.1693329278 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 13 |
Hb_006569_040 |
0.1696799031 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 14 |
Hb_018202_050 |
0.1720059295 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A [Jatropha curcas] |
| 15 |
Hb_006570_010 |
0.1727155585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001662_140 |
0.1735138713 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003900_030 |
0.1737847875 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002233_130 |
0.1762631636 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000808_110 |
0.1787175007 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 20 |
Hb_007283_010 |
0.1797958575 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |