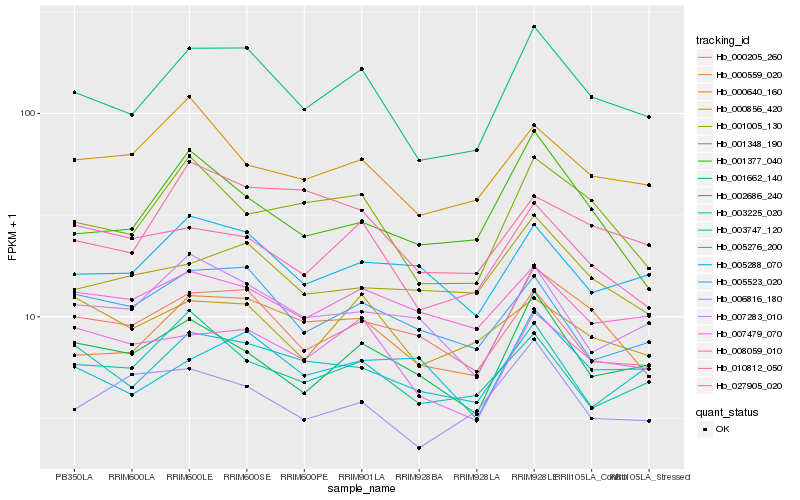
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_240 |
0.0 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 2 |
Hb_000559_020 |
0.0831016383 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 3 |
Hb_007479_070 |
0.098325498 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 4 |
Hb_001348_190 |
0.0991256418 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010812_050 |
0.1026871763 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001662_140 |
0.1038797208 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000205_260 |
0.1046149502 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 8 |
Hb_005288_070 |
0.1059951659 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_003747_120 |
0.1084659321 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005523_020 |
0.1099176662 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 11 |
Hb_008059_010 |
0.1118596226 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001005_130 |
0.1124351599 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000856_420 |
0.1134683258 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_003225_020 |
0.1140888772 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 15 |
Hb_007283_010 |
0.11428258 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 16 |
Hb_027905_020 |
0.1143467429 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 17 |
Hb_000640_160 |
0.1153938973 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 18 |
Hb_006816_180 |
0.1172408235 |
- |
- |
- |
| 19 |
Hb_005276_200 |
0.1179846748 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001377_040 |
0.118688383 |
- |
- |
chitinase, putative [Ricinus communis] |