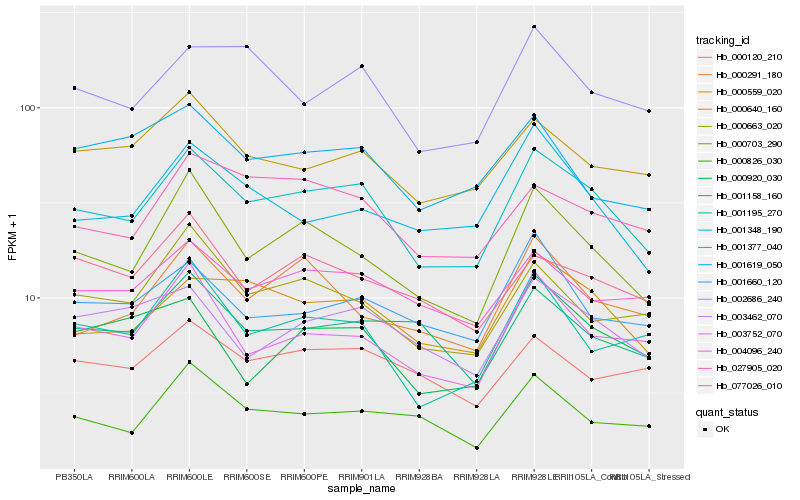
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_190 |
0.0 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000559_020 |
0.08489961 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 3 |
Hb_000703_290 |
0.0852378329 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 4 |
Hb_002686_240 |
0.0991256418 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 5 |
Hb_004096_240 |
0.0997039571 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 6 |
Hb_003752_070 |
0.100903068 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000640_160 |
0.1019705869 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 8 |
Hb_003462_070 |
0.1020921001 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 9 |
Hb_027905_020 |
0.1031799984 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 10 |
Hb_000663_020 |
0.1053395792 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001195_270 |
0.105787848 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000826_030 |
0.1079592913 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 13 |
Hb_001660_120 |
0.1096490254 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001158_160 |
0.109954908 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000120_210 |
0.111019742 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 16 |
Hb_001377_040 |
0.1118682714 |
- |
- |
chitinase, putative [Ricinus communis] |
| 17 |
Hb_077026_010 |
0.1123759995 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 18 |
Hb_000291_180 |
0.1134080361 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 19 |
Hb_000920_030 |
0.114425962 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001619_050 |
0.1151475371 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |