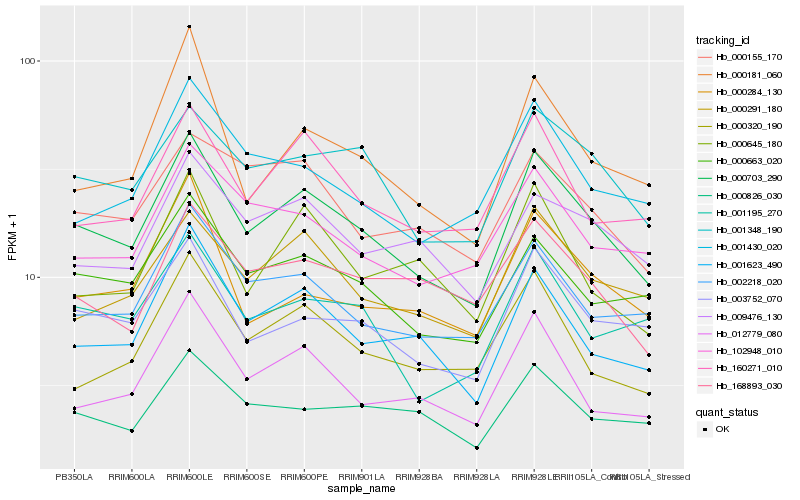
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_290 |
0.0 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 2 |
Hb_001348_190 |
0.0852378329 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003752_070 |
0.0863996495 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001623_490 |
0.0905335745 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000663_020 |
0.0907648928 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_000291_180 |
0.093293997 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 7 |
Hb_002218_020 |
0.0937684231 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 8 |
Hb_160271_010 |
0.0970849696 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001195_270 |
0.1007338773 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012779_080 |
0.1012575042 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000826_030 |
0.1013098142 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 12 |
Hb_102948_010 |
0.1035216893 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000181_060 |
0.1043573196 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 14 |
Hb_009476_130 |
0.1056695895 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 15 |
Hb_000320_190 |
0.106131465 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000155_170 |
0.1069804235 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 17 |
Hb_001430_020 |
0.1074736327 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000645_180 |
0.1086534574 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000284_130 |
0.1087307906 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 20 |
Hb_168893_030 |
0.1089593266 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |