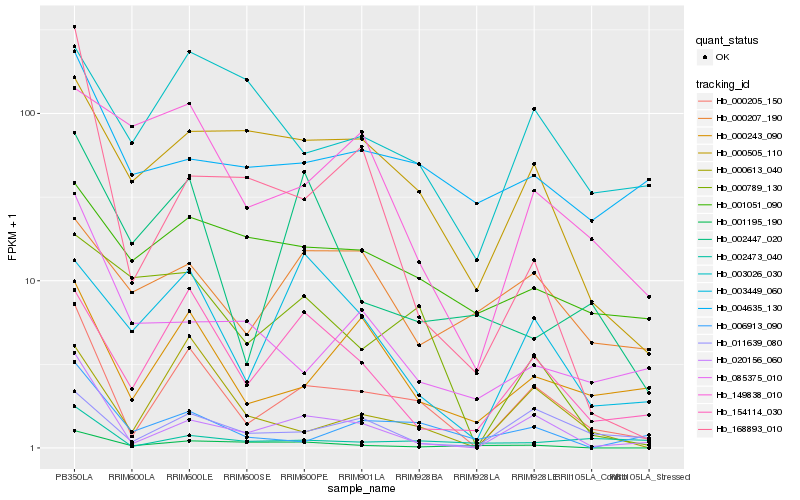
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 2 |
Hb_020156_060 |
0.1982991051 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 3 |
Hb_000243_090 |
0.2326139173 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |
| 4 |
Hb_000613_040 |
0.2346509863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011639_080 |
0.2395576338 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 6 |
Hb_154114_030 |
0.2400855414 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_006913_090 |
0.2595169244 |
- |
- |
PREDICTED: uncharacterized protein LOC101215996 [Cucumis sativus] |
| 8 |
Hb_000505_110 |
0.2649849707 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 9 |
Hb_002473_040 |
0.2711725706 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003449_060 |
0.277625988 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 11 |
Hb_004635_130 |
0.2890809332 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 12 |
Hb_003026_030 |
0.289306187 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 13 |
Hb_001195_190 |
0.2948308078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000789_130 |
0.2949900351 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 15 |
Hb_149838_010 |
0.2957699931 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 16 |
Hb_002447_020 |
0.2981707216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001051_090 |
0.2982465931 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 18 |
Hb_168893_010 |
0.3039465065 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 19 |
Hb_085375_010 |
0.3056728951 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000207_190 |
0.3093442077 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |