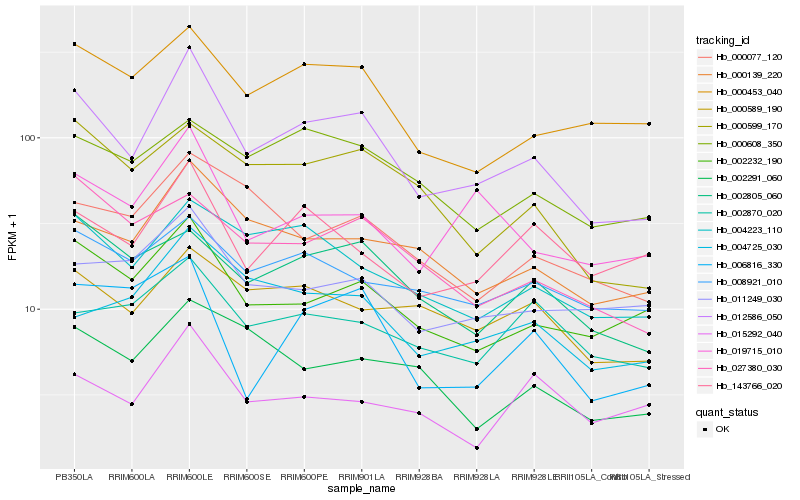
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012586_050 |
0.0 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 2 |
Hb_002232_190 |
0.1257584857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000139_220 |
0.1322725536 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000453_040 |
0.1362160933 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 5 |
Hb_004223_110 |
0.1391623838 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 6 |
Hb_019715_010 |
0.1444052 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 7 |
Hb_000599_170 |
0.1464394353 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 8 |
Hb_143766_020 |
0.1483985456 |
- |
- |
putative phospholipase A2 [Gossypium arboreum] |
| 9 |
Hb_000589_190 |
0.1494630486 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002805_060 |
0.1497432201 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 11 |
Hb_011249_030 |
0.1502119162 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 12 |
Hb_008921_010 |
0.1517653586 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 13 |
Hb_004725_030 |
0.1533727009 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 14 |
Hb_000077_120 |
0.1543809809 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 15 |
Hb_027380_030 |
0.1560289239 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_006816_330 |
0.1586065572 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 17 |
Hb_000608_350 |
0.1598789552 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 18 |
Hb_002291_060 |
0.1622339201 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_015292_040 |
0.162464407 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 20 |
Hb_002870_020 |
0.1629890489 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |