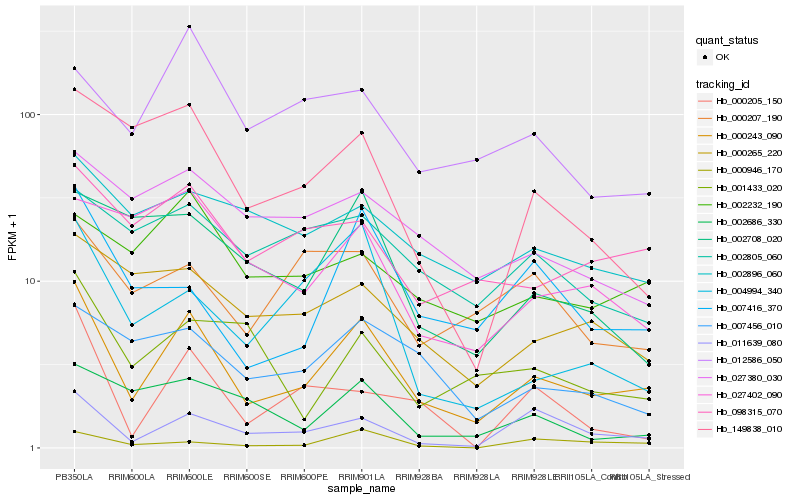
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_090 |
0.0 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |
| 2 |
Hb_011639_080 |
0.1944561064 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 3 |
Hb_007416_370 |
0.2060485976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_098315_070 |
0.2090607915 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 5 |
Hb_149838_010 |
0.2169324716 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 6 |
Hb_007456_010 |
0.2193412068 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 7 |
Hb_000265_220 |
0.2204891947 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 8 |
Hb_004994_340 |
0.2248698278 |
- |
- |
- |
| 9 |
Hb_002896_060 |
0.2265745562 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 10 |
Hb_002232_190 |
0.2266791258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_027380_030 |
0.2285090294 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000946_170 |
0.2312655642 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 13 |
Hb_002686_330 |
0.2316054558 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002805_060 |
0.2318317372 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 15 |
Hb_000207_190 |
0.2319080837 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 16 |
Hb_002708_020 |
0.232204203 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000205_150 |
0.2326139173 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 18 |
Hb_027402_090 |
0.2329505929 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 19 |
Hb_012586_050 |
0.2332098735 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 20 |
Hb_001433_020 |
0.2336956585 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |