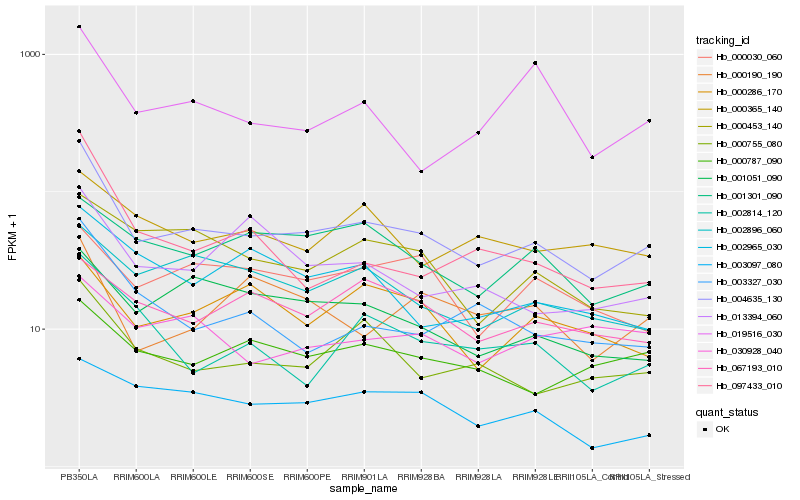
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004635_130 |
0.0 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 2 |
Hb_000755_080 |
0.1363061745 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 3 |
Hb_001051_090 |
0.156764528 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 4 |
Hb_003327_030 |
0.1568618 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_000787_090 |
0.1574380843 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 6 |
Hb_002896_060 |
0.1586560917 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 7 |
Hb_002814_120 |
0.1636347098 |
- |
- |
PREDICTED: uncharacterized protein LOC105633463 [Jatropha curcas] |
| 8 |
Hb_097433_010 |
0.1658561672 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 9 |
Hb_000190_190 |
0.1668686371 |
- |
- |
PREDICTED: BI1-like protein [Gossypium raimondii] |
| 10 |
Hb_013394_060 |
0.1675711447 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_000453_140 |
0.1677906916 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 12 |
Hb_002965_030 |
0.1679172368 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 13 |
Hb_000365_140 |
0.1684317805 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 14 |
Hb_001301_090 |
0.1686527219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_067193_010 |
0.1693058887 |
- |
- |
- |
| 16 |
Hb_003097_080 |
0.1698711095 |
- |
- |
hypothetical protein JCGZ_24481 [Jatropha curcas] |
| 17 |
Hb_019516_030 |
0.1704737654 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000030_060 |
0.1707885222 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 19 |
Hb_030928_040 |
0.1719105815 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000286_170 |
0.1720991925 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |