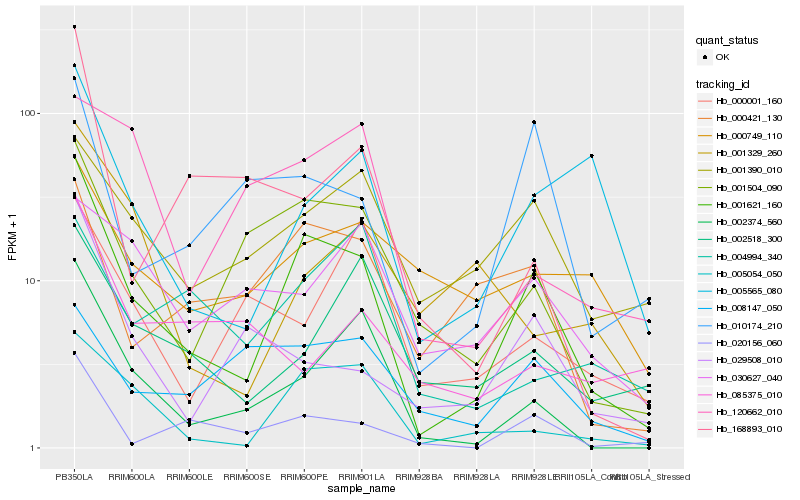
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_160 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_005054_050 |
0.219393727 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_002374_560 |
0.2252574157 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH11-like [Populus euphratica] |
| 4 |
Hb_001504_090 |
0.2289157148 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_020156_060 |
0.233875025 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 6 |
Hb_000421_130 |
0.2517969446 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_005565_080 |
0.2572134127 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 8 |
Hb_010174_210 |
0.2594425091 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001390_010 |
0.2647299148 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 10 |
Hb_002518_300 |
0.275037803 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 11 |
Hb_029508_010 |
0.2754485779 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000749_110 |
0.2835084538 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 13 |
Hb_008147_050 |
0.2900015309 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 14 |
Hb_168893_010 |
0.2944651417 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 15 |
Hb_001329_260 |
0.2989887634 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 16 |
Hb_030627_040 |
0.3008710878 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 17 |
Hb_000001_160 |
0.3033703069 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_120662_010 |
0.3093560322 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 19 |
Hb_085375_010 |
0.3128790437 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_004994_340 |
0.313247982 |
- |
- |
- |