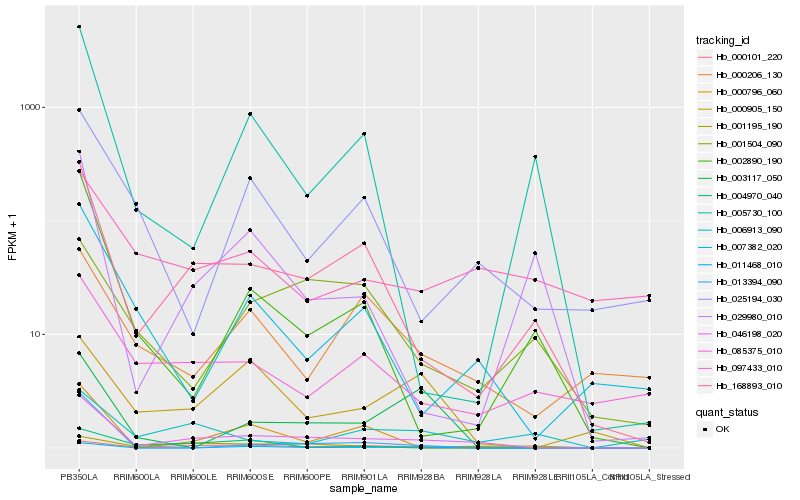
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046198_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000101_220 |
0.2076858787 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 3 |
Hb_168893_010 |
0.2107878513 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 4 |
Hb_001195_190 |
0.2717351953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011468_010 |
0.274024832 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 6 |
Hb_000796_060 |
0.2836026681 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 7 |
Hb_007382_020 |
0.2836750569 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 8 |
Hb_085375_010 |
0.2848527585 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000206_130 |
0.2991427084 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_025194_030 |
0.3001069436 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 11 |
Hb_000905_150 |
0.3010485861 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 12 |
Hb_097433_010 |
0.3101325645 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 13 |
Hb_003117_050 |
0.3101946226 |
- |
- |
- |
| 14 |
Hb_002890_190 |
0.3145258072 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 15 |
Hb_013394_090 |
0.3161919342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_029980_010 |
0.316473269 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 17 |
Hb_004970_040 |
0.3220563803 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 18 |
Hb_001504_090 |
0.3234040075 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_005730_100 |
0.3239983484 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 20 |
Hb_006913_090 |
0.3244706832 |
- |
- |
PREDICTED: uncharacterized protein LOC101215996 [Cucumis sativus] |