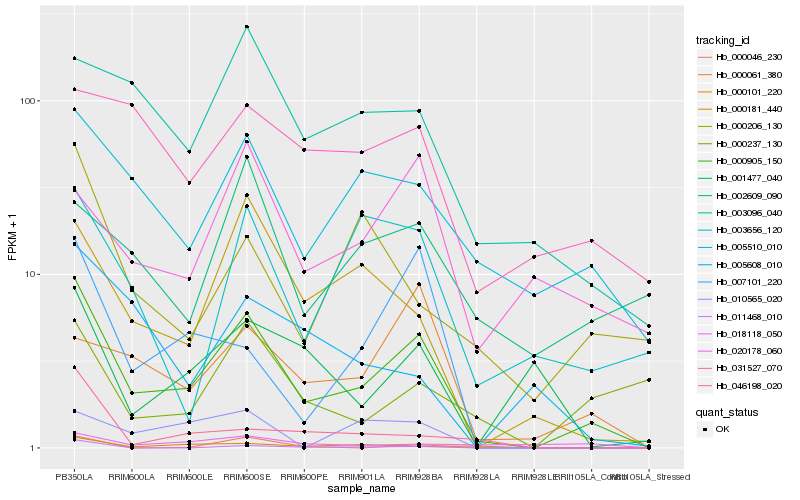
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_150 |
0.0 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 2 |
Hb_005510_010 |
0.2392779618 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000181_440 |
0.2620716616 |
- |
- |
- |
| 4 |
Hb_003096_040 |
0.2652125075 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 5 |
Hb_007101_220 |
0.2750664186 |
- |
- |
Gibberellin-regulated protein 3 precursor, putative [Ricinus communis] |
| 6 |
Hb_000101_220 |
0.2759552102 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 7 |
Hb_003656_120 |
0.2812503956 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 8 |
Hb_031527_070 |
0.2861919107 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 9 |
Hb_000061_380 |
0.2865867208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010565_020 |
0.288424692 |
- |
- |
- |
| 11 |
Hb_020178_060 |
0.2895373068 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 12 |
Hb_001477_040 |
0.2907235033 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_000206_130 |
0.2960422286 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_005608_010 |
0.2981237155 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 15 |
Hb_011468_010 |
0.298570288 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 16 |
Hb_000237_130 |
0.2986706871 |
- |
- |
- |
| 17 |
Hb_018118_050 |
0.2989985759 |
- |
- |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 18 |
Hb_002609_090 |
0.2994215268 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_000046_230 |
0.301015851 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 20 |
Hb_046198_020 |
0.3010485861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |