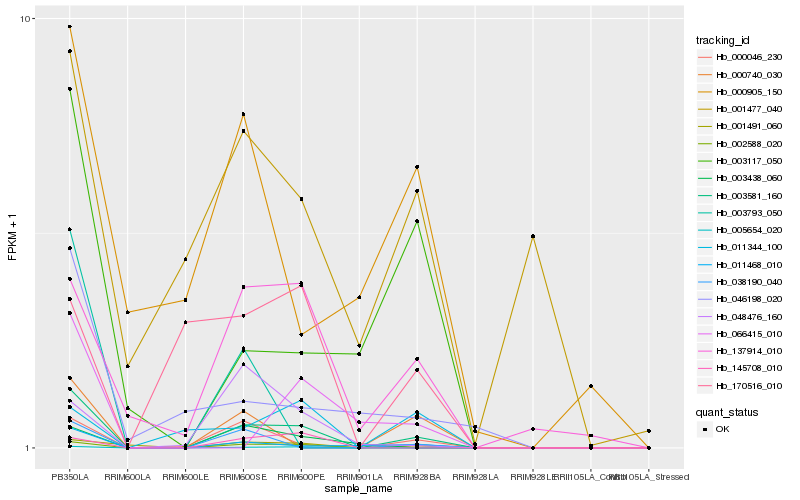
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_002588_020 |
0.163477609 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 3 |
Hb_005654_020 |
0.246312523 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_011468_010 |
0.2503585741 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 5 |
Hb_000046_230 |
0.2685114 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 6 |
Hb_001491_060 |
0.2792821887 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6-like isoform X1 [Populus euphratica] |
| 7 |
Hb_137914_010 |
0.3015371562 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 8 |
Hb_003117_050 |
0.3335189931 |
- |
- |
- |
| 9 |
Hb_145708_010 |
0.334998398 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_038190_040 |
0.3375814933 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 11 |
Hb_003438_060 |
0.3377791529 |
- |
- |
- |
| 12 |
Hb_000740_030 |
0.3405416654 |
- |
- |
hypothetical protein VITISV_038271 [Vitis vinifera] |
| 13 |
Hb_046198_020 |
0.354494712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_048476_160 |
0.3578193979 |
- |
- |
hypothetical protein JCGZ_19593 [Jatropha curcas] |
| 15 |
Hb_011344_100 |
0.3597356239 |
- |
- |
- |
| 16 |
Hb_066415_010 |
0.3766848644 |
- |
- |
- |
| 17 |
Hb_001477_040 |
0.3775379426 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 18 |
Hb_170516_010 |
0.3791449854 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 19 |
Hb_000905_150 |
0.3826212273 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 20 |
Hb_003793_050 |
0.3873684545 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |