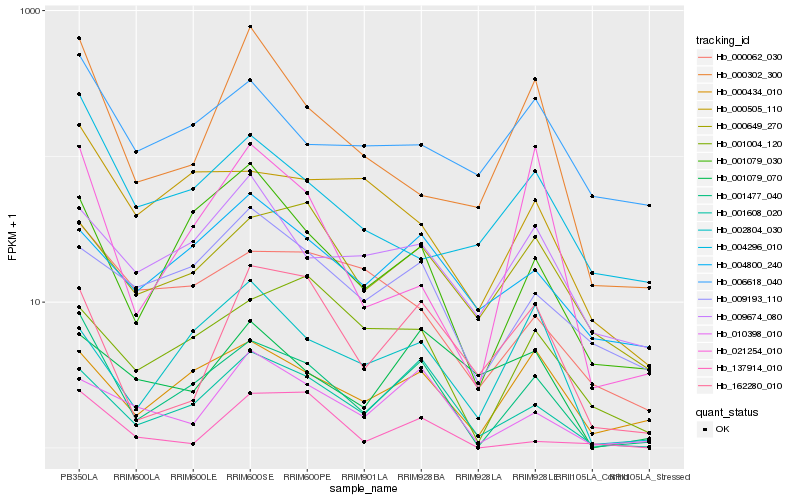
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001477_040 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 2 |
Hb_001608_020 |
0.1919496094 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001079_030 |
0.2094450363 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 4 |
Hb_000434_010 |
0.2143742305 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 5 |
Hb_010398_010 |
0.2341768131 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 6 |
Hb_000505_110 |
0.2373797604 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 7 |
Hb_000062_030 |
0.2456221212 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004296_010 |
0.2470393067 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 9 |
Hb_000649_270 |
0.2488241242 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_009193_110 |
0.2510179094 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_162280_010 |
0.2514874733 |
- |
- |
Phosphoprotein phosphatase isoform 2, partial [Theobroma cacao] |
| 12 |
Hb_006618_040 |
0.2517185415 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 13 |
Hb_000302_300 |
0.2530188407 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 14 |
Hb_021254_010 |
0.2532887848 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 15 |
Hb_001004_120 |
0.2554057777 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004800_240 |
0.2554192636 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 17 |
Hb_002804_030 |
0.2561025521 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 18 |
Hb_009674_080 |
0.2564286313 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 19 |
Hb_137914_010 |
0.2630947022 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 20 |
Hb_001079_070 |
0.2646873151 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |