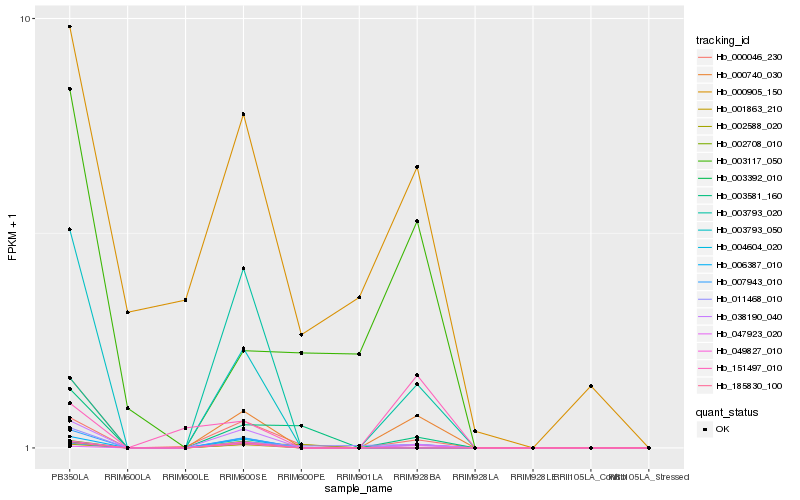
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_030 |
0.0 |
- |
- |
hypothetical protein VITISV_038271 [Vitis vinifera] |
| 2 |
Hb_038190_040 |
0.1931621272 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 3 |
Hb_000046_230 |
0.2386325228 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 4 |
Hb_004604_020 |
0.2463463188 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 5 |
Hb_011468_010 |
0.293514245 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 6 |
Hb_047923_020 |
0.2960659323 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_002588_020 |
0.3403216632 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 8 |
Hb_003581_160 |
0.3405416654 |
- |
- |
- |
| 9 |
Hb_003117_050 |
0.3426783914 |
- |
- |
- |
| 10 |
Hb_007943_010 |
0.3460711259 |
- |
- |
- |
| 11 |
Hb_000905_150 |
0.3487260452 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 12 |
Hb_003793_050 |
0.3530437944 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 13 |
Hb_151497_010 |
0.3600290241 |
- |
- |
- |
| 14 |
Hb_003392_010 |
0.3606881885 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 15 |
Hb_049827_010 |
0.3615281313 |
- |
- |
PREDICTED: uncharacterized protein LOC105762110 [Gossypium raimondii] |
| 16 |
Hb_003793_020 |
0.3616740123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006387_010 |
0.3625407123 |
- |
- |
PREDICTED: uncharacterized protein LOC105649124 [Jatropha curcas] |
| 18 |
Hb_001863_210 |
0.3650474441 |
- |
- |
- |
| 19 |
Hb_185830_100 |
0.3655064484 |
- |
- |
- |
| 20 |
Hb_002708_010 |
0.3660206857 |
- |
- |
- |