| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
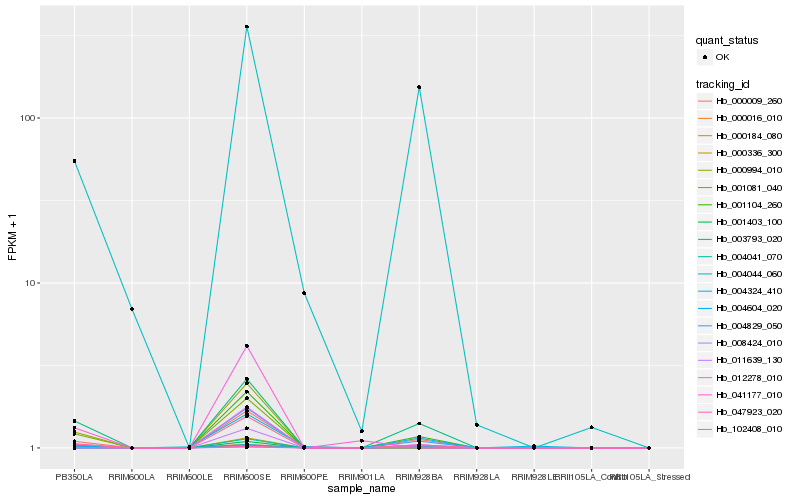
Hb_004041_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_102408_010 |
0.089365081 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 3 |
Hb_003793_020 |
0.099065839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000009_260 |
0.1131440039 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012278_010 |
0.1443656076 |
- |
- |
- |
| 6 |
Hb_004604_020 |
0.2134507747 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 7 |
Hb_004044_060 |
0.2187222521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000184_080 |
0.2283551844 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 9 |
Hb_000994_010 |
0.2314762652 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 10 |
Hb_001081_040 |
0.2321607381 |
- |
- |
hypothetical protein MIMGU_mgv11b021166mg, partial [Erythranthe guttata] |
| 11 |
Hb_047923_020 |
0.2362352618 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_000336_300 |
0.2392943393 |
- |
- |
- |
| 13 |
Hb_004324_410 |
0.2402196819 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 14 |
Hb_000016_010 |
0.2588239254 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 15 |
Hb_008424_010 |
0.259387306 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_041177_010 |
0.2717407132 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 17 |
Hb_004829_050 |
0.2733305444 |
- |
- |
hypothetical protein TRIUR3_09654 [Triticum urartu] |
| 18 |
Hb_011639_130 |
0.2733349169 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 19 |
Hb_001403_100 |
0.2734267129 |
- |
- |
hypothetical protein CICLE_v10017457mg, partial [Citrus clementina] |
| 20 |
Hb_001104_260 |
0.2734384932 |
- |
- |
hypothetical protein JCGZ_23628 [Jatropha curcas] |