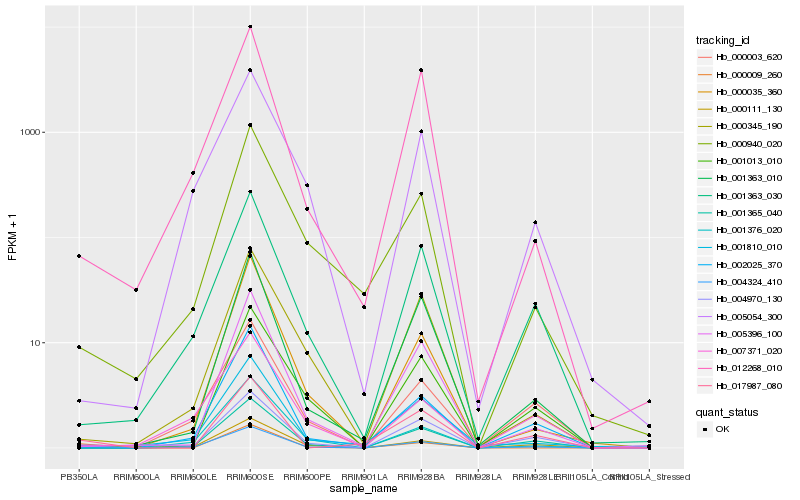
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_410 |
0.0 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 2 |
Hb_001363_030 |
0.167916119 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 3 |
Hb_004970_130 |
0.1692233569 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 4 |
Hb_001365_040 |
0.1750005271 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 5 |
Hb_000009_260 |
0.1792604659 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002025_370 |
0.1816375322 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_001810_010 |
0.1819665992 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 8 |
Hb_012268_010 |
0.1836495021 |
- |
- |
glutamic acid-rich protein [Manihot esculenta] |
| 9 |
Hb_000940_020 |
0.1843487191 |
- |
- |
hypothetical protein POPTR_0004s17500g [Populus trichocarpa] |
| 10 |
Hb_000111_130 |
0.1848897403 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 11 |
Hb_005396_100 |
0.1894580893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017987_080 |
0.1898534548 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 13 |
Hb_005054_300 |
0.1899724714 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 14 |
Hb_001363_010 |
0.1923965257 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 15 |
Hb_000345_190 |
0.1925112077 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 16 |
Hb_001376_020 |
0.1928258121 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 17 |
Hb_007371_020 |
0.1940806523 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001013_010 |
0.1943225888 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000003_620 |
0.1946485604 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 20 |
Hb_000035_360 |
0.1958953774 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |