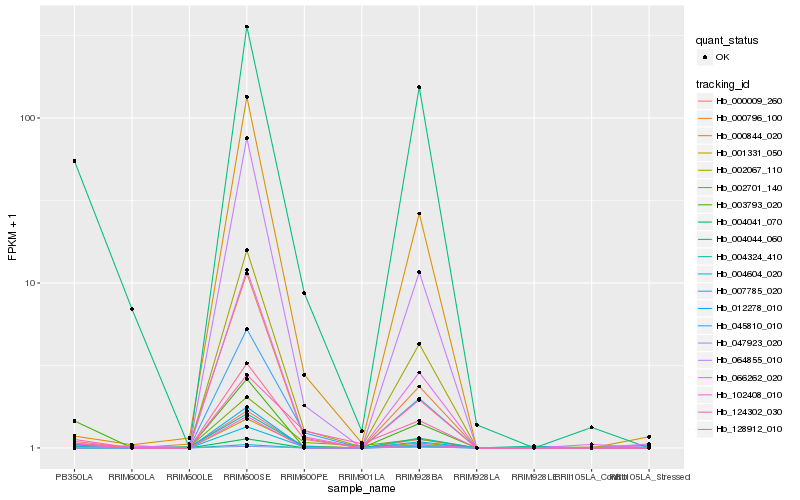
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_260 |
0.0 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004041_070 |
0.1131440039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003793_020 |
0.139885164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_102408_010 |
0.1413203488 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 5 |
Hb_004044_060 |
0.1581379076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012278_010 |
0.1788588305 |
- |
- |
- |
| 7 |
Hb_004324_410 |
0.1792604659 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 8 |
Hb_128912_010 |
0.2097620067 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 9 |
Hb_124302_030 |
0.2125619454 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000844_020 |
0.2298519119 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 11 |
Hb_000796_100 |
0.2351946157 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 12 |
Hb_047923_020 |
0.2378498584 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_002067_110 |
0.239950125 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 14 |
Hb_007785_020 |
0.2406122522 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004604_020 |
0.2407208788 |
- |
- |
PREDICTED: uncharacterized protein LOC102581016 [Solanum tuberosum] |
| 16 |
Hb_064855_010 |
0.2407217378 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_066262_020 |
0.2410862669 |
- |
- |
hypothetical protein JCGZ_14803 [Jatropha curcas] |
| 18 |
Hb_002701_140 |
0.2434266987 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 19 |
Hb_001331_050 |
0.244824333 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 20 |
Hb_045810_010 |
0.2452741559 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |