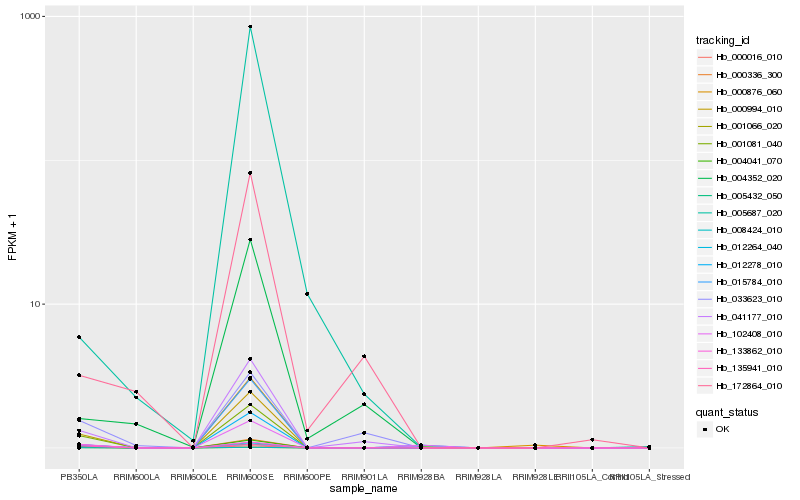
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000994_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 2 |
Hb_001081_040 |
0.0385857683 |
- |
- |
hypothetical protein MIMGU_mgv11b021166mg, partial [Erythranthe guttata] |
| 3 |
Hb_000336_300 |
0.0790949777 |
- |
- |
- |
| 4 |
Hb_000016_010 |
0.1349662673 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 5 |
Hb_008424_010 |
0.1362459925 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_041177_010 |
0.1404881012 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 7 |
Hb_005432_050 |
0.169570998 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 8 |
Hb_001066_020 |
0.1918015481 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_012278_010 |
0.1946789293 |
- |
- |
- |
| 10 |
Hb_102408_010 |
0.1979251848 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 11 |
Hb_015784_010 |
0.2041421638 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 12 |
Hb_133862_010 |
0.2073131351 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 13 |
Hb_135941_010 |
0.2105997154 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 14 |
Hb_012264_040 |
0.2220739622 |
- |
- |
- |
| 15 |
Hb_033623_010 |
0.2299667668 |
- |
- |
- |
| 16 |
Hb_004041_070 |
0.2314762652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_172864_010 |
0.2540057273 |
- |
- |
PREDICTED: 3'-hydroxy-N-methyl-(S)-coclaurine 4'-O-methyltransferase-like [Jatropha curcas] |
| 18 |
Hb_000876_060 |
0.2589849207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004352_020 |
0.2591662326 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005687_020 |
0.2625186489 |
- |
- |
P66 protein [Hevea brasiliensis] |