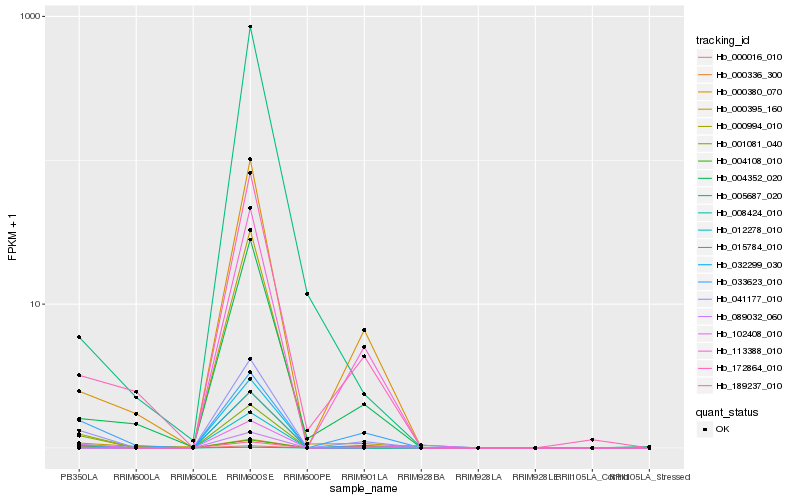
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041177_010 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 2 |
Hb_000994_010 |
0.1404881012 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 3 |
Hb_172864_010 |
0.1603795191 |
- |
- |
PREDICTED: 3'-hydroxy-N-methyl-(S)-coclaurine 4'-O-methyltransferase-like [Jatropha curcas] |
| 4 |
Hb_001081_040 |
0.1623058555 |
- |
- |
hypothetical protein MIMGU_mgv11b021166mg, partial [Erythranthe guttata] |
| 5 |
Hb_004352_020 |
0.168846806 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000380_070 |
0.1697442715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_033623_010 |
0.1737253682 |
- |
- |
- |
| 8 |
Hb_015784_010 |
0.1852631623 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 9 |
Hb_000336_300 |
0.190591062 |
- |
- |
- |
| 10 |
Hb_012278_010 |
0.206151444 |
- |
- |
- |
| 11 |
Hb_004108_010 |
0.2108898374 |
- |
- |
- |
| 12 |
Hb_089032_060 |
0.2163430116 |
- |
- |
Uncharacterized protein TCM_027342 [Theobroma cacao] |
| 13 |
Hb_032299_030 |
0.2164372311 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 14 |
Hb_189237_010 |
0.2167377649 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_102408_010 |
0.2233118908 |
- |
- |
hypothetical protein JCGZ_27060 [Jatropha curcas] |
| 16 |
Hb_000395_160 |
0.2289714476 |
- |
- |
hypothetical protein POPTR_0006s25490g [Populus trichocarpa] |
| 17 |
Hb_005687_020 |
0.229396407 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 18 |
Hb_113388_010 |
0.2332564321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000016_010 |
0.2348808665 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 20 |
Hb_008424_010 |
0.2359435082 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |