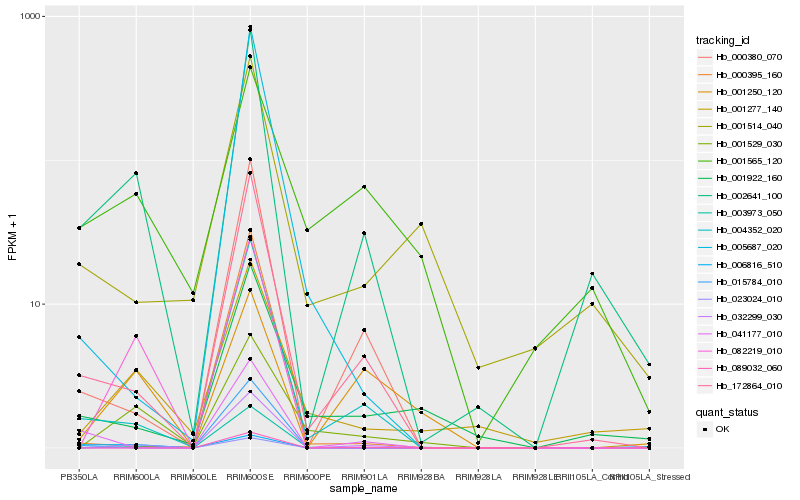
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 2 |
Hb_172864_010 |
0.1666809719 |
- |
- |
PREDICTED: 3'-hydroxy-N-methyl-(S)-coclaurine 4'-O-methyltransferase-like [Jatropha curcas] |
| 3 |
Hb_004352_020 |
0.1812496285 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001277_140 |
0.2095105319 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 5 |
Hb_000380_070 |
0.2112744362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_082219_010 |
0.222819031 |
- |
- |
Os03g0264300, partial [Oryza sativa Japonica Group] |
| 7 |
Hb_001922_160 |
0.2350064444 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_041177_010 |
0.2485529056 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 9 |
Hb_001529_030 |
0.2520216614 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 10 |
Hb_001514_040 |
0.2530579563 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 11 |
Hb_003973_050 |
0.2589161176 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 3-like isoform X1 [Populus euphratica] |
| 12 |
Hb_023024_010 |
0.2610247147 |
- |
- |
PREDICTED: uncharacterized protein LOC105767094 [Gossypium raimondii] |
| 13 |
Hb_001565_120 |
0.2624381488 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006816_510 |
0.2624918995 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 15 |
Hb_089032_060 |
0.2683749147 |
- |
- |
Uncharacterized protein TCM_027342 [Theobroma cacao] |
| 16 |
Hb_032299_030 |
0.2690862019 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 17 |
Hb_015784_010 |
0.2696343076 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 18 |
Hb_005687_020 |
0.2758937674 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 19 |
Hb_000395_160 |
0.2762635651 |
- |
- |
hypothetical protein POPTR_0006s25490g [Populus trichocarpa] |
| 20 |
Hb_001250_120 |
0.277584627 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |