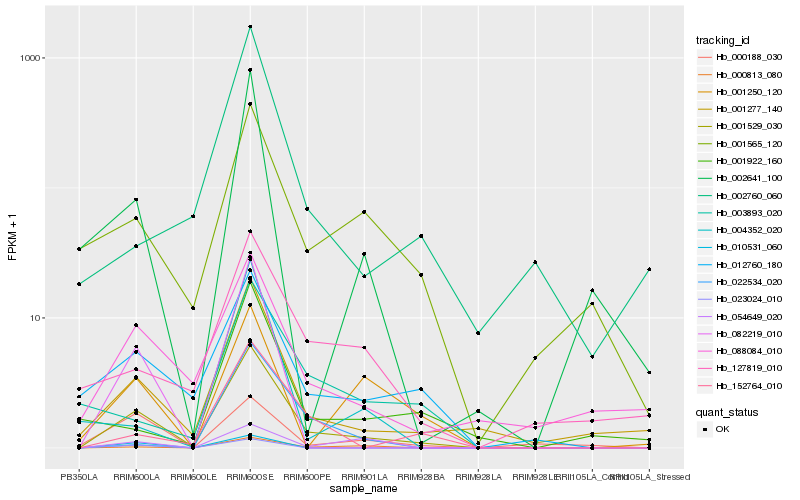
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001529_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 2 |
Hb_001277_140 |
0.1891189456 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 3 |
Hb_000813_080 |
0.2154688789 |
- |
- |
caspase, putative [Ricinus communis] |
| 4 |
Hb_082219_010 |
0.2216362876 |
- |
- |
Os03g0264300, partial [Oryza sativa Japonica Group] |
| 5 |
Hb_012760_180 |
0.2263985777 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 6 |
Hb_152764_010 |
0.2307566178 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_023024_010 |
0.2340679451 |
- |
- |
PREDICTED: uncharacterized protein LOC105767094 [Gossypium raimondii] |
| 8 |
Hb_088084_010 |
0.2421667409 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 9 |
Hb_010531_060 |
0.2518017001 |
- |
- |
hypothetical protein B456_010G209400, partial [Gossypium raimondii] |
| 10 |
Hb_002641_100 |
0.2520216614 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 11 |
Hb_001565_120 |
0.2526005608 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_127819_010 |
0.2598187146 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_022534_020 |
0.2634715184 |
- |
- |
PREDICTED: uncharacterized protein LOC103422158 [Malus domestica] |
| 14 |
Hb_001250_120 |
0.2646507419 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |
| 15 |
Hb_003893_020 |
0.2680793631 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002760_060 |
0.2731042574 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 17 |
Hb_054649_020 |
0.2741606172 |
- |
- |
PREDICTED: uncharacterized protein LOC102629934 [Citrus sinensis] |
| 18 |
Hb_001922_160 |
0.2747880261 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_000188_030 |
0.2757637919 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 20 |
Hb_004352_020 |
0.2802056727 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |