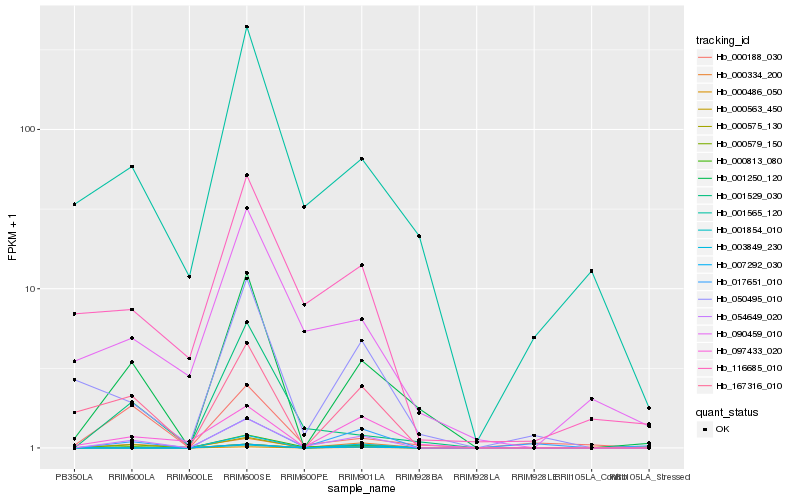
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054649_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102629934 [Citrus sinensis] |
| 2 |
Hb_000813_080 |
0.1489748672 |
- |
- |
caspase, putative [Ricinus communis] |
| 3 |
Hb_001250_120 |
0.2176861611 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |
| 4 |
Hb_017651_010 |
0.2417747241 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_167316_010 |
0.2574201637 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 1 [Nicotiana sylvestris] |
| 6 |
Hb_000579_150 |
0.2605717921 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 7 |
Hb_097433_020 |
0.2696323414 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 8 |
Hb_001529_030 |
0.2741606172 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 9 |
Hb_050495_010 |
0.2783031241 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 10 |
Hb_116685_010 |
0.2805588021 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_000334_200 |
0.2870897613 |
desease resistance |
Gene Name: RVT_1 |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_001565_120 |
0.2911784844 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000563_450 |
0.2973572976 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 14 |
Hb_000575_130 |
0.2976329398 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Populus euphratica] |
| 15 |
Hb_003849_230 |
0.2977163257 |
- |
- |
PREDICTED: NEP1-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_000486_050 |
0.2979671018 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_007292_030 |
0.3058123065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_090459_010 |
0.3101454957 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 19 |
Hb_000188_030 |
0.3195470715 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 20 |
Hb_001854_010 |
0.321068296 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |