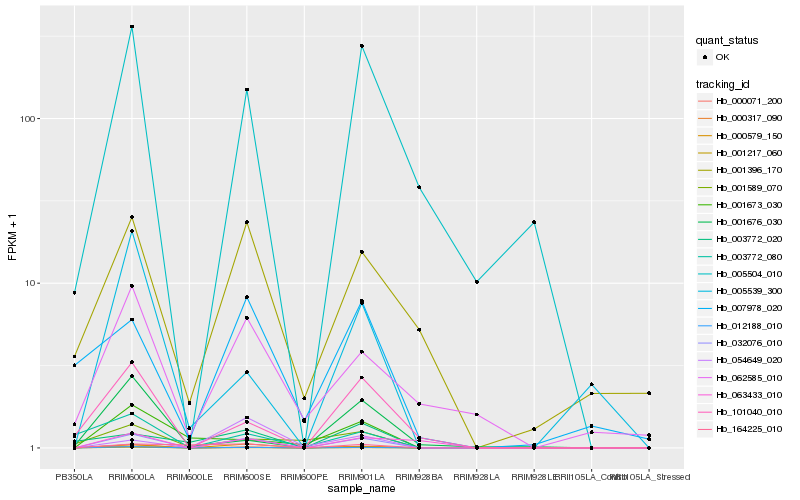
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032076_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_012188_010 |
0.0974621388 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_001217_060 |
0.1233193579 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_000071_200 |
0.1679458159 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 5 |
Hb_000579_150 |
0.1683960126 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 6 |
Hb_005504_010 |
0.2349545668 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 7 |
Hb_101040_010 |
0.2767452055 |
- |
- |
- |
| 8 |
Hb_001396_170 |
0.2965384125 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_063433_010 |
0.3031579252 |
- |
- |
Chloroplast channel forming outer membrane protein isoform 2 [Theobroma cacao] |
| 10 |
Hb_062585_010 |
0.3051600757 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |
| 11 |
Hb_007978_020 |
0.3156256496 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA1-like [Jatropha curcas] |
| 12 |
Hb_000317_090 |
0.3163027183 |
- |
- |
hypothetical protein RCOM_1176360 [Ricinus communis] |
| 13 |
Hb_003772_080 |
0.3188487065 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_005539_300 |
0.3282398729 |
- |
- |
PREDICTED: deacetoxyvindoline 4-hydroxylase-like [Populus euphratica] |
| 15 |
Hb_001676_030 |
0.3326708346 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_001673_030 |
0.3357134279 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 17 |
Hb_003772_020 |
0.3380255053 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |
| 18 |
Hb_001589_070 |
0.3383974539 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_054649_020 |
0.3408671297 |
- |
- |
PREDICTED: uncharacterized protein LOC102629934 [Citrus sinensis] |
| 20 |
Hb_164225_010 |
0.3429324639 |
- |
- |
- |