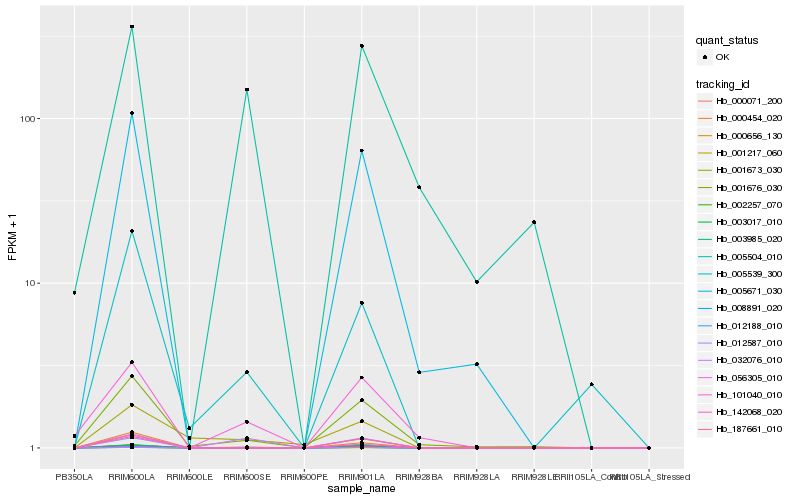
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 2 |
Hb_001217_060 |
0.1132400443 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 3 |
Hb_012188_010 |
0.1143582929 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 4 |
Hb_032076_010 |
0.1679458159 |
- |
- |
- |
| 5 |
Hb_101040_010 |
0.2078630235 |
- |
- |
- |
| 6 |
Hb_001676_030 |
0.210464488 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_005539_300 |
0.211349843 |
- |
- |
PREDICTED: deacetoxyvindoline 4-hydroxylase-like [Populus euphratica] |
| 8 |
Hb_005504_010 |
0.2354305058 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 9 |
Hb_001673_030 |
0.2636229675 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 10 |
Hb_187661_010 |
0.2752669204 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_142068_020 |
0.2832246415 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000454_020 |
0.2835942413 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_000656_130 |
0.2857646727 |
- |
- |
hypothetical protein PRUPE_ppa026157mg, partial [Prunus persica] |
| 14 |
Hb_005671_030 |
0.2888571132 |
- |
- |
- |
| 15 |
Hb_008891_020 |
0.2897100964 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 16 |
Hb_056305_010 |
0.2897391636 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_002257_070 |
0.2898633752 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 18 |
Hb_003017_010 |
0.2902208375 |
- |
- |
PREDICTED: uncharacterized protein LOC104878882 [Vitis vinifera] |
| 19 |
Hb_003985_020 |
0.2912260917 |
- |
- |
- |
| 20 |
Hb_012587_010 |
0.292725812 |
- |
- |
- |