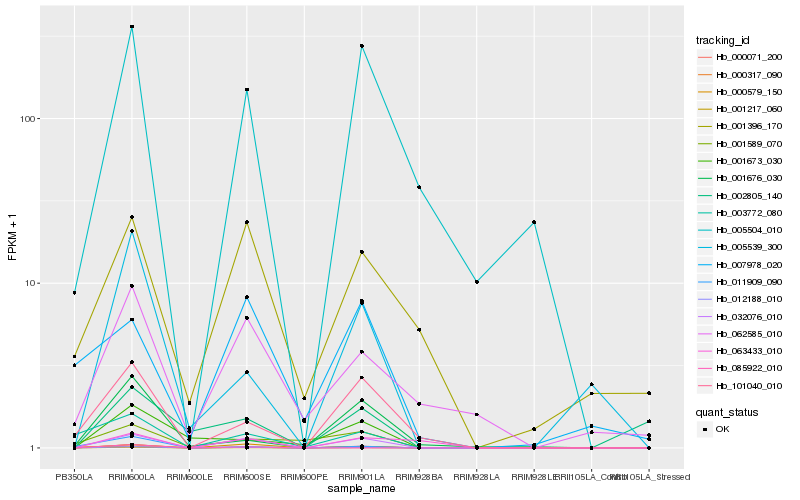
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012188_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 2 |
Hb_001217_060 |
0.0260909956 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 3 |
Hb_032076_010 |
0.0974621388 |
- |
- |
- |
| 4 |
Hb_000071_200 |
0.1143582929 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 5 |
Hb_000579_150 |
0.1976907956 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 6 |
Hb_005504_010 |
0.2428477638 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 7 |
Hb_101040_010 |
0.2639050846 |
- |
- |
- |
| 8 |
Hb_005539_300 |
0.2695584166 |
- |
- |
PREDICTED: deacetoxyvindoline 4-hydroxylase-like [Populus euphratica] |
| 9 |
Hb_062585_010 |
0.2915639434 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |
| 10 |
Hb_001676_030 |
0.2950848966 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_003772_080 |
0.2988117425 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 12 |
Hb_001673_030 |
0.3095702615 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 13 |
Hb_063433_010 |
0.3127988064 |
- |
- |
Chloroplast channel forming outer membrane protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_001396_170 |
0.3146317556 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_011909_090 |
0.3240383462 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105633212 [Jatropha curcas] |
| 16 |
Hb_000317_090 |
0.3344992313 |
- |
- |
hypothetical protein RCOM_1176360 [Ricinus communis] |
| 17 |
Hb_001589_070 |
0.3345255191 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 18 |
Hb_002805_140 |
0.3458497343 |
- |
- |
hypothetical protein POPTR_0001s28390g [Populus trichocarpa] |
| 19 |
Hb_085922_010 |
0.3558259244 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_007978_020 |
0.3643791739 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA1-like [Jatropha curcas] |