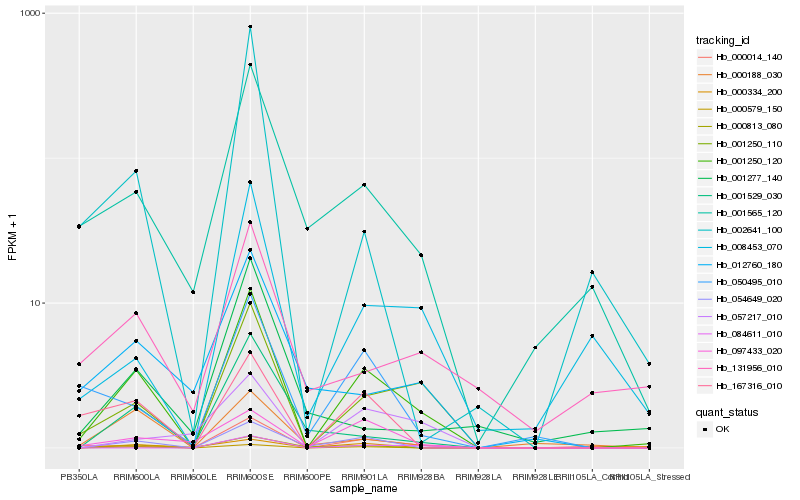
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001250_120 |
0.0 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |
| 2 |
Hb_001250_110 |
0.1767543955 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0010s05340g [Populus trichocarpa] |
| 3 |
Hb_054649_020 |
0.2176861611 |
- |
- |
PREDICTED: uncharacterized protein LOC102629934 [Citrus sinensis] |
| 4 |
Hb_000014_140 |
0.237840233 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 5 |
Hb_001565_120 |
0.254105194 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000813_080 |
0.2541313334 |
- |
- |
caspase, putative [Ricinus communis] |
| 7 |
Hb_167316_010 |
0.2587520342 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 1 [Nicotiana sylvestris] |
| 8 |
Hb_008453_070 |
0.262909192 |
- |
- |
putative aquaporin [Vitis vinifera] |
| 9 |
Hb_001529_030 |
0.2646507419 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 10 |
Hb_050495_010 |
0.2697902209 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_097433_020 |
0.2742711769 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 12 |
Hb_002641_100 |
0.277584627 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 13 |
Hb_000188_030 |
0.2840225687 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 14 |
Hb_012760_180 |
0.2852348029 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 15 |
Hb_000579_150 |
0.2886736028 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 16 |
Hb_057217_010 |
0.2893260892 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 17 |
Hb_131956_010 |
0.3009785697 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 18 |
Hb_084611_010 |
0.3028707398 |
- |
- |
PREDICTED: uncharacterized protein LOC104249812 [Nicotiana sylvestris] |
| 19 |
Hb_000334_200 |
0.3034142997 |
desease resistance |
Gene Name: RVT_1 |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_001277_140 |
0.3091547002 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |