| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
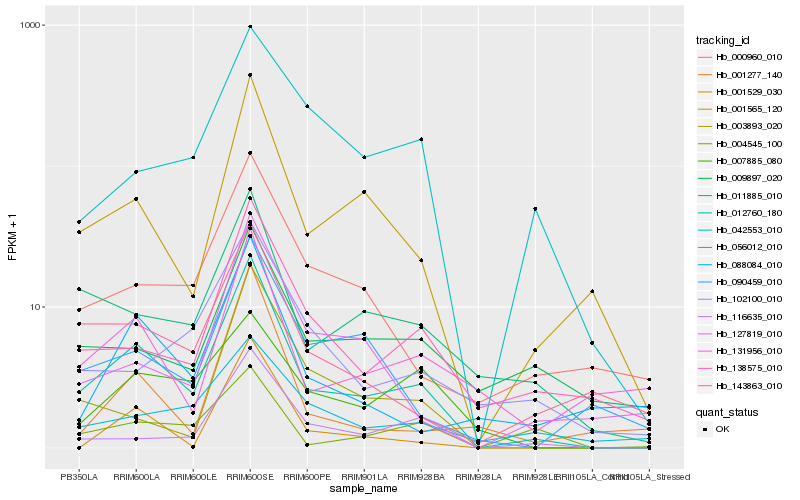
| 1 |
Hb_012760_180 |
0.0 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 2 |
Hb_001565_120 |
0.1691348507 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011885_010 |
0.1934197951 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_138575_010 |
0.2007120767 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_116635_010 |
0.2010204083 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_143863_010 |
0.2078529159 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 7 |
Hb_000960_010 |
0.2109534507 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_131956_010 |
0.218757935 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 9 |
Hb_090459_010 |
0.2210036952 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 10 |
Hb_127819_010 |
0.2253918858 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_003893_020 |
0.2263222551 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001529_030 |
0.2263985777 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 13 |
Hb_102100_010 |
0.2274574038 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 14 |
Hb_088084_010 |
0.2276999016 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 15 |
Hb_004545_100 |
0.2301873911 |
- |
- |
PREDICTED: protein BONZAI 3 [Jatropha curcas] |
| 16 |
Hb_042553_010 |
0.2313632831 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 17 |
Hb_056012_010 |
0.2380695912 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 18 |
Hb_007885_080 |
0.239794653 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001277_140 |
0.2399227064 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 20 |
Hb_009897_020 |
0.2424347995 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |