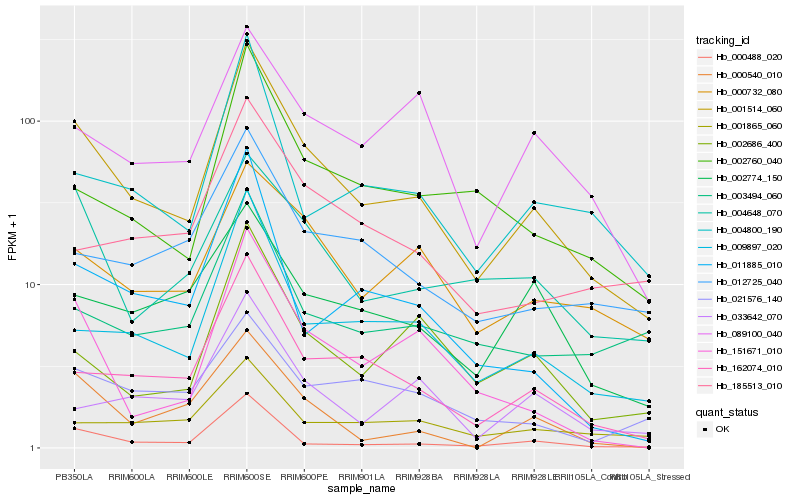
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_060 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 2 |
Hb_162074_010 |
0.1480231859 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_009897_020 |
0.1533080541 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_000488_020 |
0.1657629653 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_004648_070 |
0.1676254318 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000732_080 |
0.1714805182 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 7 |
Hb_002760_040 |
0.1719871713 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 8 |
Hb_151671_010 |
0.1731522542 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 9 |
Hb_004800_190 |
0.1754890662 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 10 |
Hb_012725_040 |
0.1806750216 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_002686_400 |
0.1812477799 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 12 |
Hb_021576_140 |
0.1864613117 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 13 |
Hb_003494_060 |
0.1867942571 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000540_010 |
0.1875688488 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 15 |
Hb_001865_060 |
0.1889713724 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 16 |
Hb_185513_010 |
0.1899596281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011885_010 |
0.1907366386 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_089100_040 |
0.1912361578 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 19 |
Hb_002774_150 |
0.191864312 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 20 |
Hb_033642_070 |
0.1941753454 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |