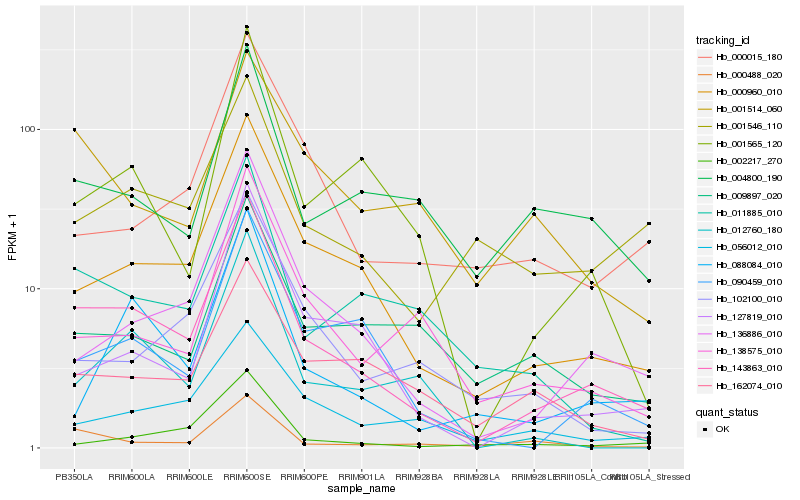
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143863_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 2 |
Hb_000960_010 |
0.1431962691 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_136886_010 |
0.179463105 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_090459_010 |
0.1848286732 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 5 |
Hb_001565_120 |
0.1979157261 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000488_020 |
0.1993064231 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 7 |
Hb_011885_010 |
0.2023857284 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_001546_110 |
0.2042974183 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 3 [Jatropha curcas] |
| 9 |
Hb_088084_010 |
0.2051416122 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 10 |
Hb_056012_010 |
0.20557693 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 11 |
Hb_127819_010 |
0.2056654023 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_012760_180 |
0.2078529159 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 13 |
Hb_138575_010 |
0.2102437842 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_004800_190 |
0.2126122675 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 15 |
Hb_162074_010 |
0.2127985538 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_102100_010 |
0.2160350257 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_002217_270 |
0.2170114966 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 18 |
Hb_001514_060 |
0.217409578 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 19 |
Hb_000015_180 |
0.2187154299 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 20 |
Hb_009897_020 |
0.2242041086 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |