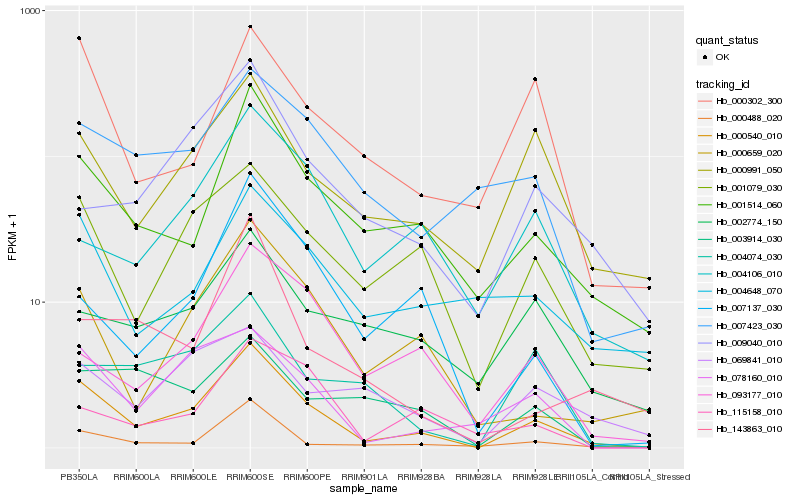
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_010 |
0.0 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 2 |
Hb_001514_060 |
0.1875688488 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 3 |
Hb_001079_030 |
0.1988966322 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 4 |
Hb_078160_010 |
0.2102601639 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 5 |
Hb_000659_020 |
0.2107703315 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 6 |
Hb_000488_020 |
0.211189807 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 7 |
Hb_000991_050 |
0.2137503223 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 8 |
Hb_000302_300 |
0.2309897043 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 9 |
Hb_007423_030 |
0.2322180992 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 10 |
Hb_004648_070 |
0.235544747 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_007137_030 |
0.2366725146 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 12 |
Hb_093177_010 |
0.2371552065 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 13 |
Hb_004074_030 |
0.2435161878 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_004106_010 |
0.2445089766 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009040_010 |
0.2508348891 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_002774_150 |
0.252990335 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 17 |
Hb_115158_010 |
0.2531607683 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_069841_010 |
0.2535017695 |
- |
- |
resistance protein [Quercus suber] |
| 19 |
Hb_143863_010 |
0.2538591232 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 20 |
Hb_003914_030 |
0.2549521167 |
- |
- |
- |