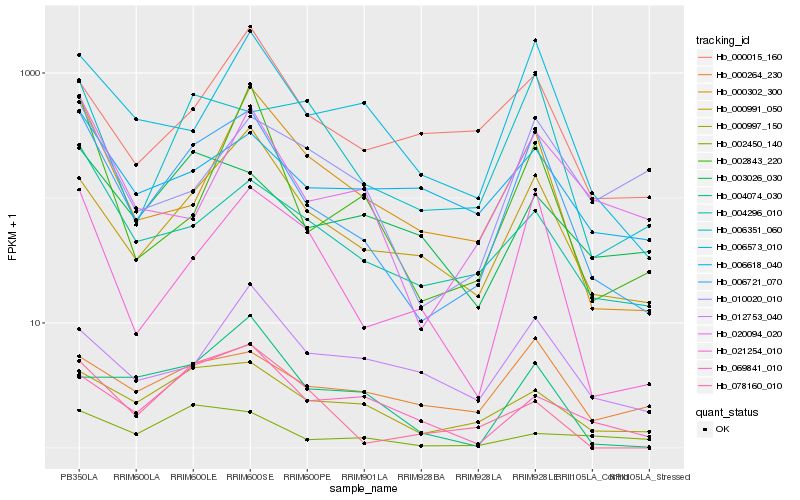
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006721_070 |
0.0 |
- |
- |
PREDICTED: 70 kDa peptidyl-prolyl isomerase-like [Jatropha curcas] |
| 2 |
Hb_000991_050 |
0.1882443021 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 3 |
Hb_021254_010 |
0.1882726701 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 4 |
Hb_000302_300 |
0.2048802803 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 5 |
Hb_006573_010 |
0.206265453 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 6 |
Hb_000997_150 |
0.2178044077 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_004296_010 |
0.2178659272 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 8 |
Hb_002843_220 |
0.2200607767 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020094_020 |
0.225006732 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 10 |
Hb_078160_010 |
0.2311915314 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 11 |
Hb_006351_060 |
0.2373066202 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 12 |
Hb_069841_010 |
0.2408982623 |
- |
- |
resistance protein [Quercus suber] |
| 13 |
Hb_000264_230 |
0.2439941472 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 14 |
Hb_006618_040 |
0.2443729804 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 15 |
Hb_000015_160 |
0.2463542996 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 16 |
Hb_012753_040 |
0.2523247648 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010020_010 |
0.2541351903 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 18 |
Hb_003026_030 |
0.2564674267 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 19 |
Hb_004074_030 |
0.2575939389 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_002450_140 |
0.2589333796 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |