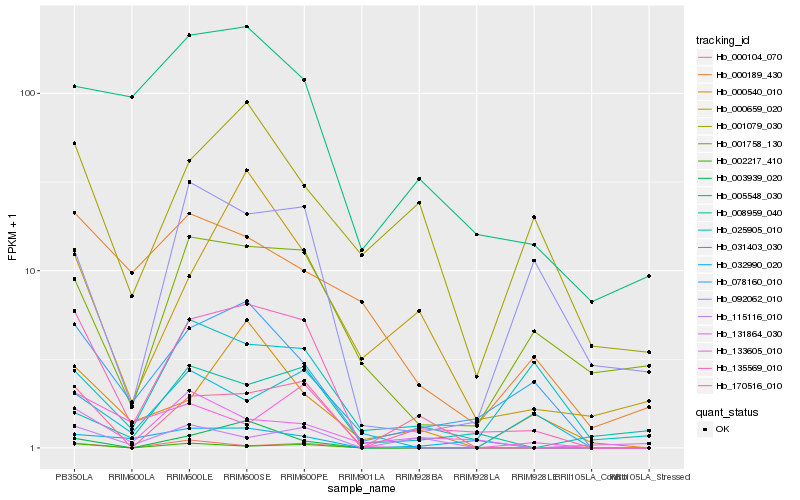
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135569_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 2 |
Hb_078160_010 |
0.2005654955 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 3 |
Hb_170516_010 |
0.2330199891 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 4 |
Hb_002217_410 |
0.2452540504 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 5 |
Hb_005548_030 |
0.2603602713 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 6 |
Hb_032990_020 |
0.2656558968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_115116_010 |
0.2681290078 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_003939_020 |
0.2690835182 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 9 |
Hb_001758_130 |
0.2773117146 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 10 |
Hb_000104_070 |
0.2779921616 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 11 |
Hb_000659_020 |
0.2834352991 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 12 |
Hb_092062_010 |
0.2846671019 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 13 |
Hb_000540_010 |
0.2850365085 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 14 |
Hb_131864_030 |
0.2852920902 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_133605_010 |
0.2882471609 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 16 |
Hb_031403_030 |
0.2906644311 |
- |
- |
- |
| 17 |
Hb_008959_040 |
0.2946380907 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 18 |
Hb_001079_030 |
0.2968615986 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 19 |
Hb_000189_430 |
0.2998719273 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_025905_010 |
0.2999618115 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |