| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
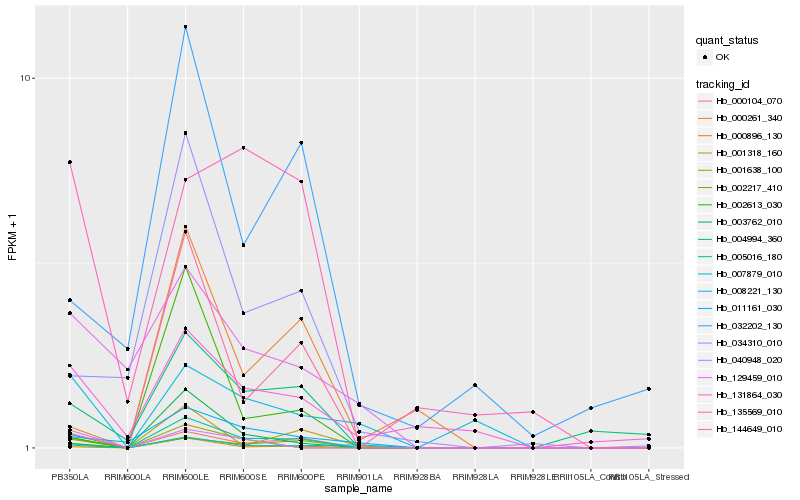
Hb_005016_180 |
0.0 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 2 |
Hb_000104_070 |
0.1278719183 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 3 |
Hb_002217_410 |
0.1740167634 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 4 |
Hb_000896_130 |
0.2130918603 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 5 |
Hb_004994_360 |
0.2496752591 |
- |
- |
PREDICTED: putative ripening-related protein 1 [Populus euphratica] |
| 6 |
Hb_003762_010 |
0.2647335318 |
- |
- |
- |
| 7 |
Hb_011161_030 |
0.2657314051 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 8 |
Hb_001638_100 |
0.2706329525 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 [Vitis vinifera] |
| 9 |
Hb_034310_010 |
0.2721354418 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_007879_010 |
0.2830213319 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 11 |
Hb_144649_010 |
0.2942235207 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 12 |
Hb_131864_030 |
0.2972666751 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_129459_010 |
0.3017341173 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 14 |
Hb_135569_010 |
0.3019262792 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 15 |
Hb_002613_030 |
0.3062326711 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_040948_020 |
0.3068616851 |
- |
- |
- |
| 17 |
Hb_032202_130 |
0.3070675432 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 18 |
Hb_001318_160 |
0.3121037849 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_000261_340 |
0.31818191 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_008221_130 |
0.3188217634 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |