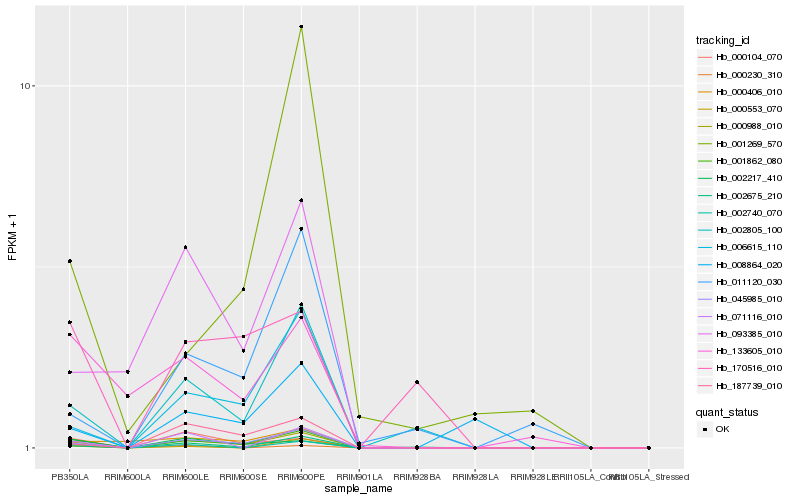
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_080 |
0.0 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 2 |
Hb_045985_010 |
0.1106528623 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 3 |
Hb_000988_010 |
0.2047589028 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 4 |
Hb_002217_410 |
0.2097718828 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 5 |
Hb_002805_100 |
0.2143744595 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 6 |
Hb_006615_110 |
0.2149017543 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 7 |
Hb_002675_210 |
0.2260777298 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 8 |
Hb_002740_070 |
0.2371103514 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 9 |
Hb_000104_070 |
0.2434151908 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 10 |
Hb_187739_010 |
0.2598963692 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_133605_010 |
0.2676856753 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_071116_010 |
0.2776627329 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 13 |
Hb_000553_070 |
0.2780206761 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000406_010 |
0.2809452577 |
- |
- |
PREDICTED: uncharacterized protein LOC105647846 [Jatropha curcas] |
| 15 |
Hb_093385_010 |
0.2811361138 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 16 |
Hb_170516_010 |
0.2862631638 |
- |
- |
hypothetical protein MTR_001s0023 [Medicago truncatula] |
| 17 |
Hb_011120_030 |
0.2954766757 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 18 |
Hb_000230_310 |
0.3007569559 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_001269_570 |
0.3031400503 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 20 |
Hb_008864_020 |
0.3045112025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |