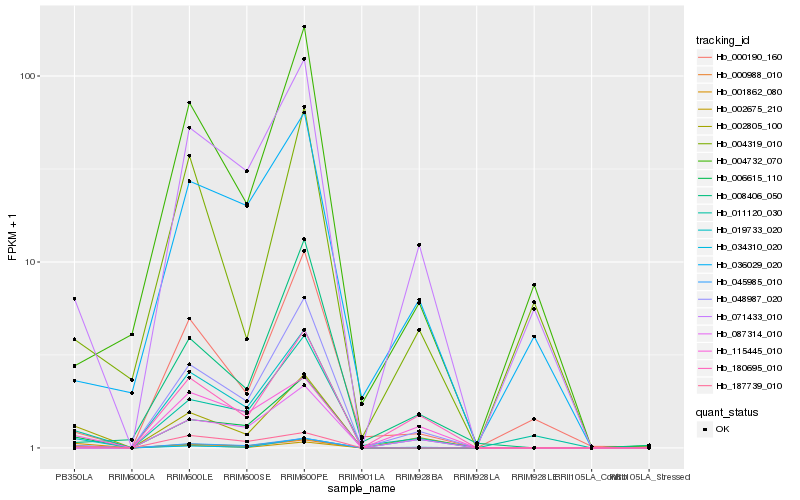
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002805_100 |
0.0 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 2 |
Hb_002675_210 |
0.059655964 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 3 |
Hb_045985_010 |
0.1792396333 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 4 |
Hb_000988_010 |
0.1916273605 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 5 |
Hb_019733_020 |
0.1964787595 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 6 |
Hb_011120_030 |
0.1998048615 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 7 |
Hb_115445_010 |
0.2022501729 |
- |
- |
- |
| 8 |
Hb_006615_110 |
0.2043937563 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 9 |
Hb_071433_010 |
0.2059727084 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001862_080 |
0.2143744595 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 11 |
Hb_180695_010 |
0.252228026 |
- |
- |
hypothetical protein CICLE_v10013449mg [Citrus clementina] |
| 12 |
Hb_000190_160 |
0.2556501899 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 13 |
Hb_048987_020 |
0.2558679976 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 14 |
Hb_008406_050 |
0.2565739169 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_004319_010 |
0.2619075109 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 16 |
Hb_187739_010 |
0.2625741911 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 17 |
Hb_087314_010 |
0.2625762554 |
- |
- |
- |
| 18 |
Hb_004732_070 |
0.2658845487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_036029_020 |
0.2676194398 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 20 |
Hb_034310_020 |
0.2678963214 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Jatropha curcas] |