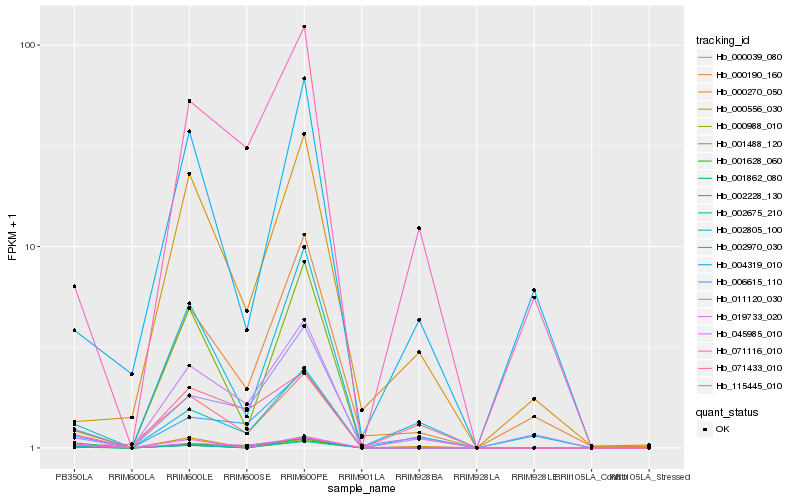
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_210 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 2 |
Hb_002805_100 |
0.059655964 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 3 |
Hb_000988_010 |
0.1836299908 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 4 |
Hb_045985_010 |
0.1852072419 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 5 |
Hb_019733_020 |
0.1911406186 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 6 |
Hb_006615_110 |
0.2119688259 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 7 |
Hb_011120_030 |
0.213521862 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 8 |
Hb_071433_010 |
0.2202701999 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001628_060 |
0.2239683417 |
- |
- |
PREDICTED: beta-galactosidase 13-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001862_080 |
0.2260777298 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 11 |
Hb_115445_010 |
0.2271739367 |
- |
- |
- |
| 12 |
Hb_001488_120 |
0.2304731174 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 13 |
Hb_004319_010 |
0.2318293225 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 14 |
Hb_002228_130 |
0.2325019348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_071116_010 |
0.2347286167 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 16 |
Hb_000190_160 |
0.237077873 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000556_030 |
0.241167394 |
- |
- |
PREDICTED: probable carboxylesterase 9 [Jatropha curcas] |
| 18 |
Hb_000270_050 |
0.2479243087 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 19 |
Hb_002970_030 |
0.2484824447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000039_080 |
0.2488666327 |
- |
- |
cyclin d, putative [Ricinus communis] |