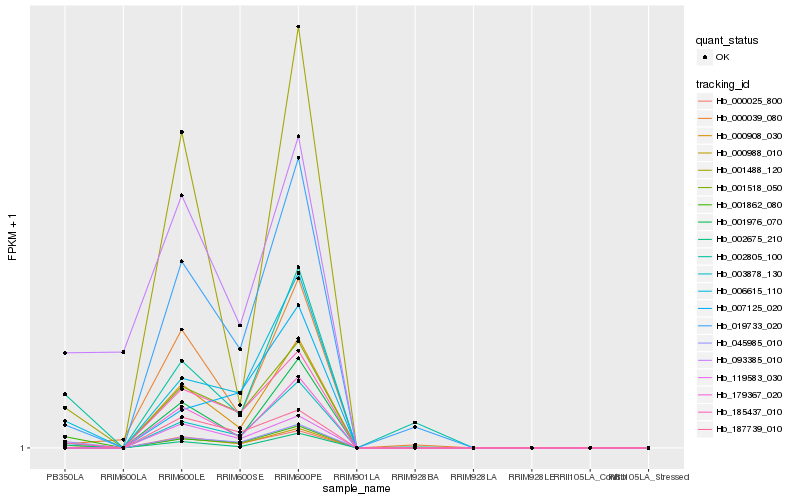
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000988_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 2 |
Hb_006615_110 |
0.0858633331 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 3 |
Hb_045985_010 |
0.0983281031 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 4 |
Hb_187739_010 |
0.1196884373 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_019733_020 |
0.1379679795 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 6 |
Hb_002675_210 |
0.1836299908 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 7 |
Hb_001518_050 |
0.191451857 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_002805_100 |
0.1916273605 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 9 |
Hb_000039_080 |
0.1919911378 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 10 |
Hb_000908_030 |
0.1932830995 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_003878_130 |
0.1946330354 |
- |
- |
- |
| 12 |
Hb_000025_800 |
0.1955113138 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 13 |
Hb_185437_010 |
0.1957816909 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 14 |
Hb_179367_020 |
0.1973168314 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_001488_120 |
0.2003298691 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 16 |
Hb_007125_020 |
0.2037544875 |
- |
- |
hypothetical protein POPTR_0003s18010g [Populus trichocarpa] |
| 17 |
Hb_001862_080 |
0.2047589028 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 18 |
Hb_093385_010 |
0.2048810055 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 19 |
Hb_119583_030 |
0.2048987533 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001976_070 |
0.2055110795 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |