| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
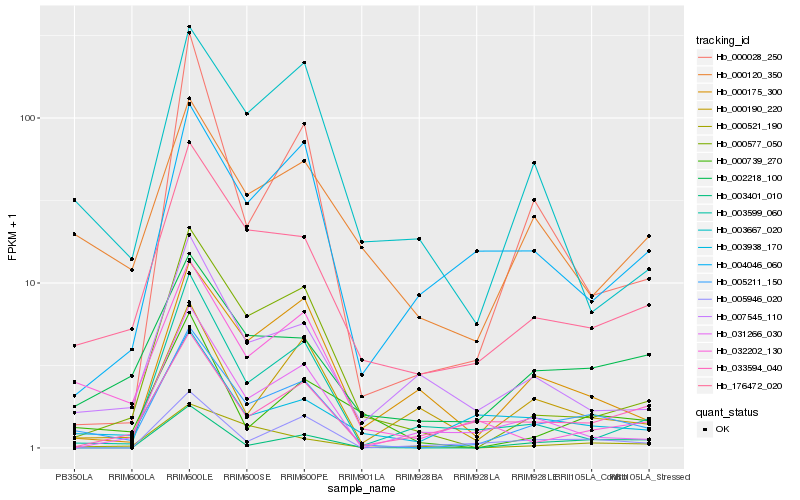
Hb_005211_150 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38) isoform X1 [Jatropha curcas] |
| 2 |
Hb_176472_020 |
0.1642402823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002218_100 |
0.1951292009 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 4 |
Hb_000577_050 |
0.2052842005 |
- |
- |
- |
| 5 |
Hb_003938_170 |
0.2168973826 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 6 |
Hb_007545_110 |
0.2179658313 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000175_300 |
0.2185868273 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 8 |
Hb_000120_350 |
0.2196047156 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 9 |
Hb_032202_130 |
0.2237668699 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 10 |
Hb_000739_270 |
0.223828039 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 11 |
Hb_003667_020 |
0.2240661202 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 12 |
Hb_000190_220 |
0.2262861361 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 13 |
Hb_005946_020 |
0.2265373439 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |
| 14 |
Hb_003599_060 |
0.2266015512 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_000028_250 |
0.2284632808 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 16 |
Hb_033594_040 |
0.2308115787 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_031266_030 |
0.2319719028 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_004046_060 |
0.2320592328 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 19 |
Hb_003401_010 |
0.2322694253 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 20 |
Hb_000521_190 |
0.232532927 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |