| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
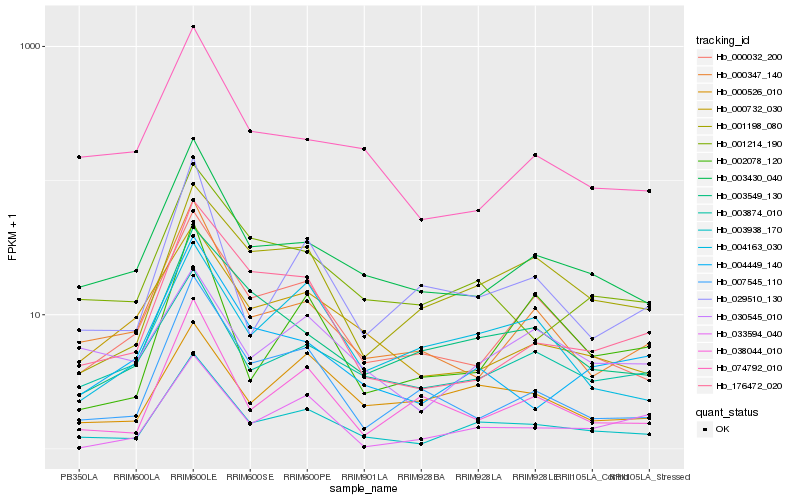
Hb_003938_170 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 2 |
Hb_003430_040 |
0.1383298082 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 3 |
Hb_003874_010 |
0.1584330341 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000032_200 |
0.1597318808 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 5 |
Hb_001198_080 |
0.1640500215 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001214_190 |
0.1649025336 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 7 |
Hb_000347_140 |
0.1652534151 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_029510_130 |
0.1659544915 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 9 |
Hb_004449_140 |
0.1679016776 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 10 |
Hb_176472_020 |
0.1706379386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003549_130 |
0.1725598342 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_074792_010 |
0.1750443474 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 13 |
Hb_000732_030 |
0.1780211296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_038044_010 |
0.181146829 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 15 |
Hb_007545_110 |
0.181501111 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_002078_120 |
0.1838273182 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 17 |
Hb_000526_010 |
0.1857502413 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 18 |
Hb_033594_040 |
0.1858923925 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_030545_010 |
0.1862965908 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 20 |
Hb_004163_030 |
0.1874094893 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |