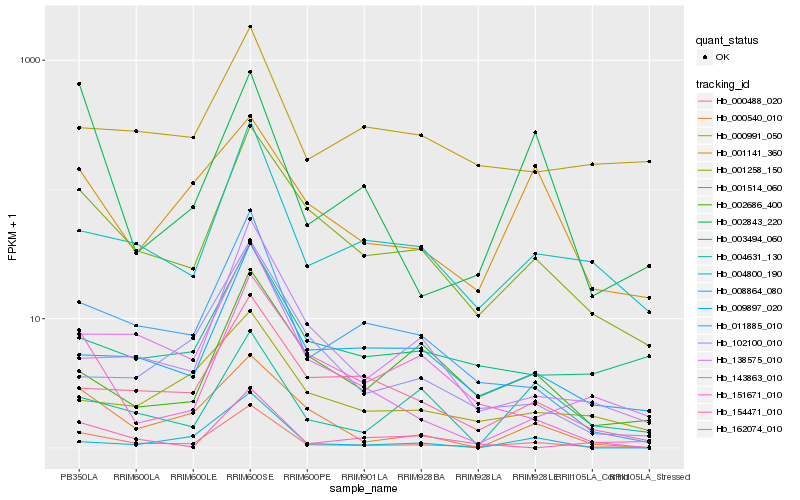
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000488_020 |
0.0 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_001514_060 |
0.1657629653 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 3 |
Hb_011885_010 |
0.1709223328 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_004800_190 |
0.1837938632 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 5 |
Hb_143863_010 |
0.1993064231 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 6 |
Hb_009897_020 |
0.205541643 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 7 |
Hb_000540_010 |
0.211189807 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 8 |
Hb_162074_010 |
0.2181462743 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 9 |
Hb_138575_010 |
0.2283348401 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_008864_080 |
0.2291942401 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 11 |
Hb_003494_060 |
0.2407178201 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 12 |
Hb_151671_010 |
0.2422623147 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 13 |
Hb_001258_150 |
0.2433746254 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 14 |
Hb_102100_010 |
0.2440246625 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_002686_400 |
0.2445116486 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 16 |
Hb_000991_050 |
0.2459236832 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 17 |
Hb_002843_220 |
0.2463104996 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001141_360 |
0.2469925146 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 19 |
Hb_004631_130 |
0.248633998 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_154471_010 |
0.2487066089 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |