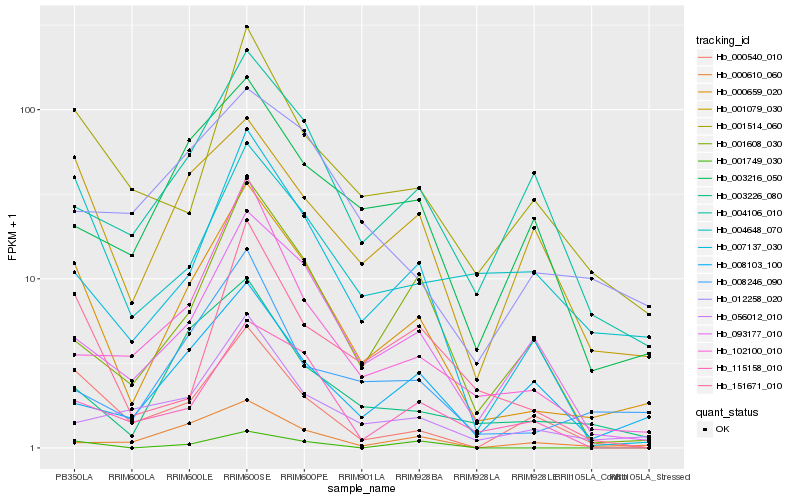
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000659_020 |
0.0 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 2 |
Hb_007137_030 |
0.1671858876 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 3 |
Hb_003226_080 |
0.1964703276 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 4 |
Hb_001079_030 |
0.1985014668 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 5 |
Hb_093177_010 |
0.1997440718 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 6 |
Hb_001514_060 |
0.2011978837 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 7 |
Hb_008246_090 |
0.2050810391 |
- |
- |
- |
| 8 |
Hb_151671_010 |
0.2096394822 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 9 |
Hb_000540_010 |
0.2107703315 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 10 |
Hb_001608_030 |
0.2179986842 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 11 |
Hb_102100_010 |
0.2200087088 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_115158_010 |
0.2203686187 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_004106_010 |
0.2224619059 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003216_050 |
0.2244571746 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 15 |
Hb_056012_010 |
0.2258198252 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 16 |
Hb_001749_030 |
0.2325507227 |
- |
- |
- |
| 17 |
Hb_004648_070 |
0.2331917849 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_008103_100 |
0.2337957587 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 19 |
Hb_000610_060 |
0.2339085722 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 20 |
Hb_012258_020 |
0.2345911186 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |