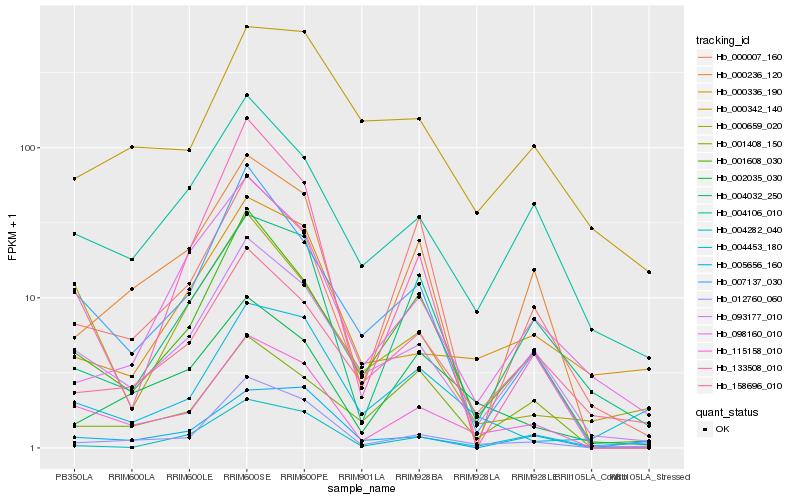
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115158_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_093177_010 |
0.1310474087 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 3 |
Hb_001608_030 |
0.1614885641 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 4 |
Hb_007137_030 |
0.1615007852 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 5 |
Hb_012760_060 |
0.1621578787 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004106_010 |
0.1698526543 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000236_120 |
0.1796437778 |
- |
- |
- |
| 8 |
Hb_158696_010 |
0.1914338102 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 9 |
Hb_002035_030 |
0.2005026188 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 10 |
Hb_004032_250 |
0.2010638679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_133508_010 |
0.2142416235 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 12 |
Hb_000336_190 |
0.2147642746 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 13 |
Hb_001408_150 |
0.21564065 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004453_180 |
0.2188387363 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 15 |
Hb_000659_020 |
0.2203686187 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 16 |
Hb_004282_040 |
0.2205856795 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000342_140 |
0.2214451982 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 18 |
Hb_098160_010 |
0.2220784324 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000007_160 |
0.2222022015 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 20 |
Hb_005656_160 |
0.2224939156 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |