| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
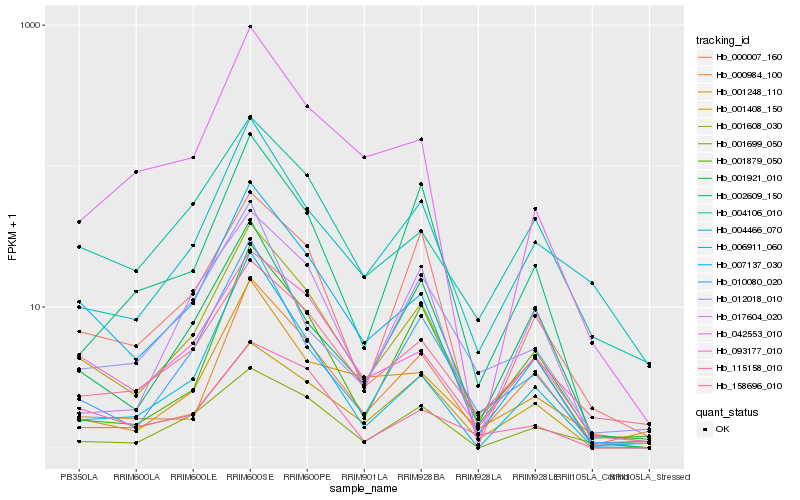
Hb_001608_030 |
0.0 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 2 |
Hb_007137_030 |
0.1048124221 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 3 |
Hb_093177_010 |
0.1207131534 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_158696_010 |
0.1315255826 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 5 |
Hb_010080_020 |
0.1321495301 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 6 |
Hb_000007_160 |
0.1415447492 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 7 |
Hb_000984_100 |
0.1439115976 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_001408_150 |
0.1465570556 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004106_010 |
0.1482088654 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004466_070 |
0.1500128251 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 11 |
Hb_001248_110 |
0.152666011 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_001879_050 |
0.1527584361 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 13 |
Hb_002609_150 |
0.1535234509 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001921_010 |
0.1540093805 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_042553_010 |
0.1554051108 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 16 |
Hb_001699_050 |
0.1585418636 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 17 |
Hb_017604_020 |
0.1607444758 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 18 |
Hb_115158_010 |
0.1614885641 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 19 |
Hb_012018_010 |
0.1653534056 |
- |
- |
hypothetical protein MIMGU_mgv1a0024251mg, partial [Erythranthe guttata] |
| 20 |
Hb_006911_060 |
0.1663540402 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |