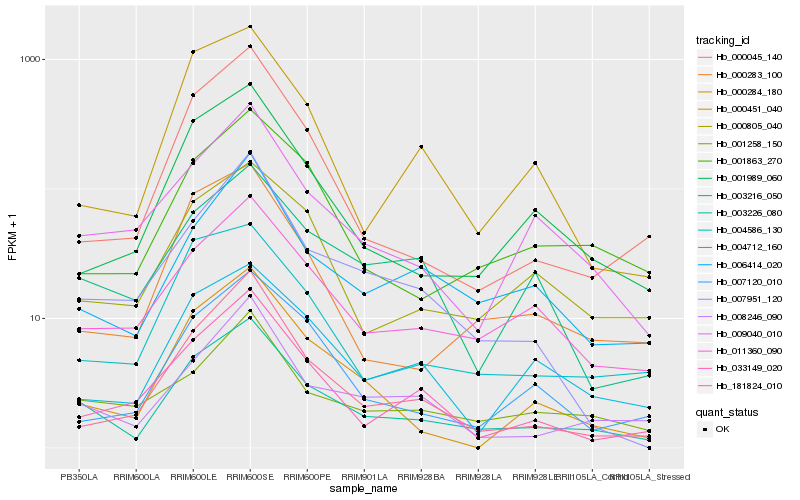
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_080 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 2 |
Hb_011360_090 |
0.1418640318 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 3 |
Hb_008246_090 |
0.1455234107 |
- |
- |
- |
| 4 |
Hb_004586_130 |
0.1488704274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006414_020 |
0.1504027403 |
transcription factor |
TF Family: MYB |
- |
| 6 |
Hb_001989_060 |
0.1528231483 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 7 |
Hb_009040_010 |
0.1588087595 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001258_150 |
0.1593561999 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 9 |
Hb_000283_100 |
0.16131113 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000805_040 |
0.161865872 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 11 |
Hb_001863_270 |
0.164930098 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 12 |
Hb_000284_180 |
0.1656352026 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 13 |
Hb_004712_160 |
0.1662634729 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 14 |
Hb_007951_120 |
0.1665600889 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 15 |
Hb_181824_010 |
0.1665797137 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 16 |
Hb_000451_040 |
0.1678817126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000045_140 |
0.170324359 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 18 |
Hb_003216_050 |
0.1708540099 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 19 |
Hb_033149_020 |
0.1730794418 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 20 |
Hb_007120_010 |
0.1783858488 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |