| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
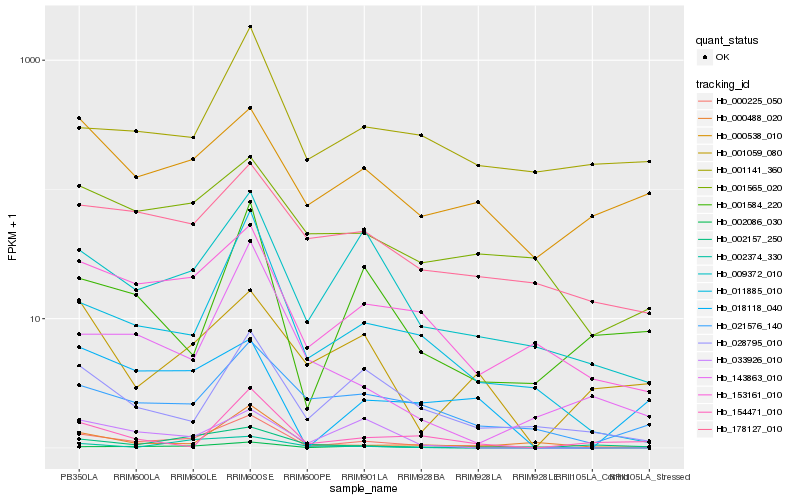
Hb_000225_050 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_011885_010 |
0.1905424652 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_009372_010 |
0.213438584 |
- |
- |
- |
| 4 |
Hb_153161_010 |
0.217633193 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 5 |
Hb_021576_140 |
0.2403201839 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 6 |
Hb_028795_010 |
0.2522124649 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_018118_040 |
0.2526427507 |
- |
- |
- |
| 8 |
Hb_000488_020 |
0.2527722336 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_143863_010 |
0.2546510977 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 10 |
Hb_002086_030 |
0.2561540995 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_001565_020 |
0.2583829377 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 12 |
Hb_033926_010 |
0.2623241058 |
- |
- |
- |
| 13 |
Hb_001584_220 |
0.2625625121 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 14 |
Hb_178127_010 |
0.2636761007 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 15 |
Hb_001141_360 |
0.2682902156 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 16 |
Hb_001059_080 |
0.2689924046 |
- |
- |
PREDICTED: F-box protein At1g61340-like [Musa acuminata subsp. malaccensis] |
| 17 |
Hb_000538_010 |
0.2706998157 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002374_330 |
0.2709584979 |
- |
- |
- |
| 19 |
Hb_002157_250 |
0.2716563598 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 20 |
Hb_154471_010 |
0.2724039386 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |