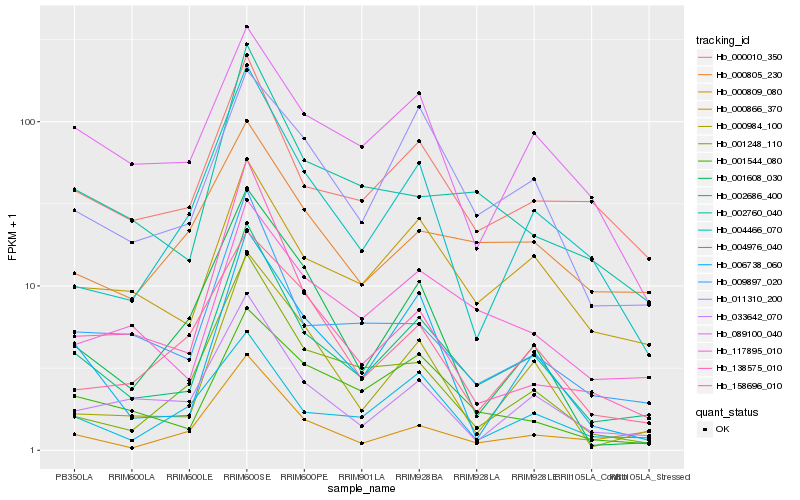
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_400 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 2 |
Hb_009897_020 |
0.1365802447 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_000010_350 |
0.1450302499 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 4 |
Hb_033642_070 |
0.1453625343 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 5 |
Hb_002760_040 |
0.1461969029 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 6 |
Hb_000866_370 |
0.1468362813 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 7 |
Hb_006738_060 |
0.1495257718 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_004466_070 |
0.1569866153 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 9 |
Hb_011310_200 |
0.1594025439 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 10 |
Hb_117895_010 |
0.1603197846 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 11 |
Hb_000984_100 |
0.1603798423 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001248_110 |
0.1634352438 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_004976_040 |
0.1651099406 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 14 |
Hb_001544_080 |
0.1652246997 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 15 |
Hb_089100_040 |
0.1697985225 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 16 |
Hb_000809_080 |
0.1723410688 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 17 |
Hb_001608_030 |
0.1737322102 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 18 |
Hb_000805_230 |
0.1780437392 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 19 |
Hb_158696_010 |
0.1781008591 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 20 |
Hb_138575_010 |
0.1781299803 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |