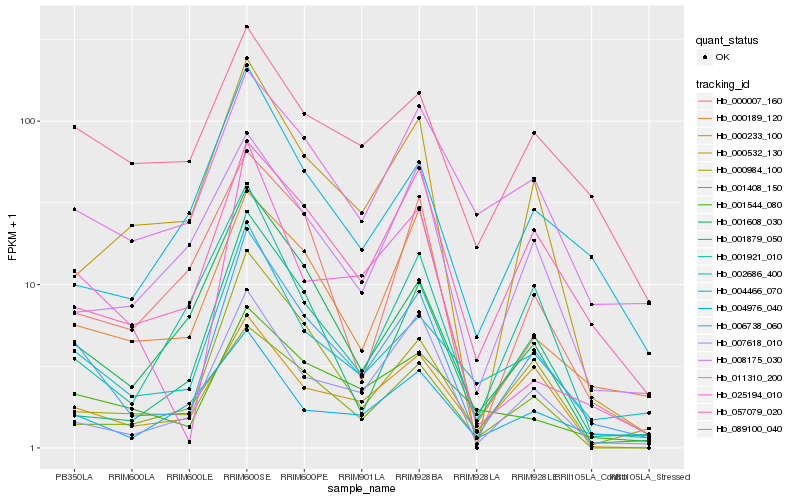
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006738_060 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002686_400 |
0.1495257718 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 3 |
Hb_000984_100 |
0.159691847 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_057079_020 |
0.1622570711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007618_010 |
0.1692747206 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 6 |
Hb_000189_120 |
0.1714952495 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 7 |
Hb_001608_030 |
0.1741375214 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 8 |
Hb_000532_130 |
0.1768746769 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 9 |
Hb_001879_050 |
0.1801946068 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 10 |
Hb_000233_100 |
0.1821826033 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 11 |
Hb_011310_200 |
0.1829801129 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 12 |
Hb_004466_070 |
0.1855480859 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 13 |
Hb_001544_080 |
0.1886028987 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 14 |
Hb_089100_040 |
0.1904970936 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 15 |
Hb_025194_010 |
0.1909215551 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 16 |
Hb_001408_150 |
0.1909467995 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001921_010 |
0.1910630505 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_008175_030 |
0.1913452634 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_004976_040 |
0.1926236466 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 20 |
Hb_000007_160 |
0.1940580946 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |