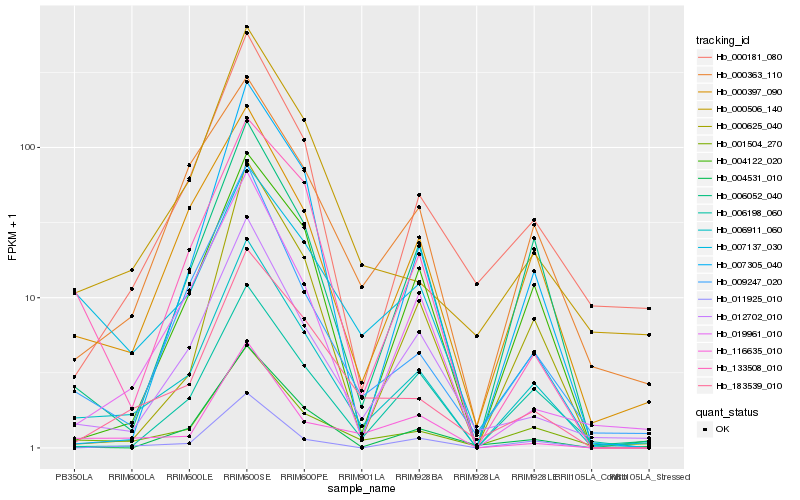
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006911_060 |
0.0 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 2 |
Hb_001504_270 |
0.1156934816 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 3 |
Hb_000397_090 |
0.1234297536 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 4 |
Hb_006052_040 |
0.1375473565 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 5 |
Hb_012702_010 |
0.1442484922 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 6 |
Hb_009247_020 |
0.1446607351 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 7 |
Hb_183539_010 |
0.1471060492 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 8 |
Hb_004122_020 |
0.1483205846 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 9 |
Hb_000181_080 |
0.1511095016 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 10 |
Hb_006198_060 |
0.1515414278 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 11 |
Hb_000363_110 |
0.1530797566 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 12 |
Hb_011925_010 |
0.1545059219 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004531_010 |
0.1581569184 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 14 |
Hb_007305_040 |
0.1585437143 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 15 |
Hb_133508_010 |
0.1609586097 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 16 |
Hb_000625_040 |
0.161200853 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 17 |
Hb_000506_140 |
0.1616054092 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 18 |
Hb_019961_010 |
0.1635333857 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 19 |
Hb_007137_030 |
0.1655157667 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 20 |
Hb_116635_010 |
0.1660590203 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |