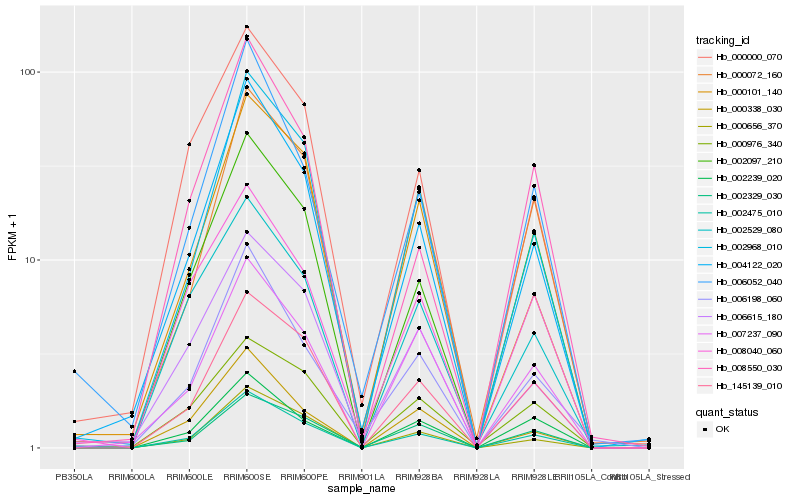
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002475_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 2 |
Hb_004122_020 |
0.093832742 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 3 |
Hb_145139_010 |
0.0990874953 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |
| 4 |
Hb_002968_010 |
0.101505959 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 5 |
Hb_006052_040 |
0.1054705814 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 6 |
Hb_002097_210 |
0.1090196168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000656_370 |
0.1120034654 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000000_070 |
0.1171425479 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 9 |
Hb_008040_060 |
0.1200603698 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 10 |
Hb_006615_180 |
0.1223082944 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 11 |
Hb_000072_160 |
0.1238242509 |
- |
- |
proline oxidase [Manihot esculenta] |
| 12 |
Hb_000976_340 |
0.1261639057 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002239_020 |
0.1294821829 |
- |
- |
MLO, putative [Theobroma cacao] |
| 14 |
Hb_002529_080 |
0.1298828187 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 15 |
Hb_006198_060 |
0.1300200722 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 16 |
Hb_002329_030 |
0.1302968815 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_007237_090 |
0.132907485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008550_030 |
0.1341495293 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 19 |
Hb_000101_140 |
0.135577004 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000338_030 |
0.1362524498 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |